NtrBC Regulates Invasiveness and Virulence of Pseudomonas aeruginosa During High-Density Infection
- PMID: 32431676
- PMCID: PMC7214821
- DOI: 10.3389/fmicb.2020.00773
NtrBC Regulates Invasiveness and Virulence of Pseudomonas aeruginosa During High-Density Infection
Abstract
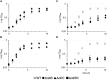
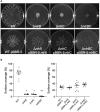
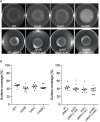
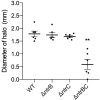
Pseudomonas aeruginosa is an opportunistic pathogen that is a major cause of nosocomial and chronic infections contributing to morbidity and mortality in cystic fibrosis patients. One of the reasons for its success as a pathogen is its ability to adapt to a broad range of circumstances. Here, we show the involvement of the general nitrogen regulator NtrBC, which is structurally conserved but functionally diverse across species, in pathogenic and adaptive states of P. aeruginosa. The role of NtrB and NtrC was examined in progressive or chronic infections, which revealed that mutants (ΔntrB, ΔntrC, and ΔntrBC) were reduced in their ability to invade and cause damage in a high-density abscess model in vivo. Progressive infections were established with mutants in the highly virulent PA14 genetic background, whereas chronic infections were established with mutants in the less virulent clinical isolate LESB58 genetic background. Characterization of adaptive lifestyles in vitro confirmed that the double ΔntrBC mutant demonstrated >40% inhibition of biofilm formation, a nearly complete inhibition of swarming motility, and a modest decrease and altered surfing motility colony appearance; with the exception of swarming, single mutants generally had more subtle or no changes. Transcriptional profiles of deletion mutants under swarming conditions were defined using RNA-Seq and unveiled dysregulated expression of hundreds of genes implicated in virulence in PA14 and LESB58 chronic lung infections, as well as carbon and nitrogen metabolism. Thus, transcriptional profiles were validated by testing responsiveness of mutants to several key intermediates of central metabolic pathways. These results indicate that NtrBC is a global regulatory system involved in both pathological and physiological processes relevant to the success of Pseudomonas in high-density infection.
Keywords: NtrC; Pseudomonas aeruginosa; abscess; adaptive lifestyles; high-density infection; invasiveness; nitrogen metabolism; virulence.
Copyright © 2020 Alford, Baghela, Yeung, Pletzer and Hancock.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

