Genotyping-by-sequencing illuminates high levels of divergence among sympatric forms of coregonines in the Laurentian Great Lakes
- PMID: 32431751
- PMCID: PMC7232772
- DOI: 10.1111/eva.12919
Genotyping-by-sequencing illuminates high levels of divergence among sympatric forms of coregonines in the Laurentian Great Lakes
Abstract
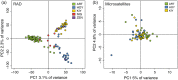
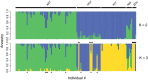
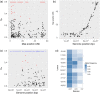
Effective resource management depends on our ability to partition diversity into biologically meaningful units. Recent evolutionary divergence, however, can often lead to ambiguity in morphological and genetic differentiation, complicating the delineation of valid conservation units. Such is the case with the "coregonine problem," where recent postglacial radiations of coregonines into lacustrine habitats resulted in the evolution of numerous species flocks, often with ambiguous taxonomy. The application of genomics methods is beginning to shed light on this problem and the evolutionary mechanisms underlying divergence in these ecologically and economically important fishes. Here, we used restriction site-associated DNA (RAD) sequencing to examine genetic diversity and differentiation among sympatric forms in the Coregonus artedi complex in the Apostle Islands of Lake Superior, the largest lake in the Laurentian Great Lakes. Using 29,068 SNPs, we were able to clearly distinguish among the three most common forms for the first time, as well as identify putative hybrids and potentially misidentified specimens. Population assignment rates for these forms using our RAD data were 93%-100% with the only mis-assignments arising from putative hybrids, an improvement from 62% to 77% using microsatellites. Estimates of pairwise differentiation (F ST: 0.045-0.056) were large given the detection of hybrids, suggesting that reduced fitness of hybrid individuals may be a potential mechanism for the maintenance of differentiation. We also used a newly built C. artedi linkage map to look for islands of genetic divergence among forms and found widespread differentiation across the genome, a pattern indicative of long-term drift, suggesting that these forms have been reproductively isolated for a substantial amount of time. The results of this study provide valuable information that can be applied to develop well-informed management strategies and stress the importance of re-evaluating conservation units with genomic tools to ensure they accurately reflect species diversity.
Keywords: RAD sequencing; adaptive divergence; conservation units; coregonines; genomic islands of divergence; hybridization; population genomics; species complex.
© 2020 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Allendorf, F. , & Luikart, G. (2007). Conservation and the genetics of populations. Oxford, UK:Blackwell Publishing Ltd.
Associated data
LinkOut - more resources
Full Text Sources

