Construction and Validation of an Autophagy-Related Prognostic Risk Signature for Survival Predicting in Clear Cell Renal Cell Carcinoma Patients
- PMID: 32432045
- PMCID: PMC7214632
- DOI: 10.3389/fonc.2020.00707
Construction and Validation of an Autophagy-Related Prognostic Risk Signature for Survival Predicting in Clear Cell Renal Cell Carcinoma Patients
Abstract
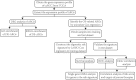
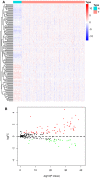
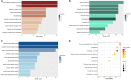
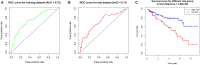
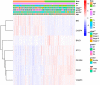
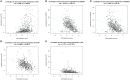
Background: Clear cell renal cell carcinoma (ccRCC) is a common type of malignant tumors in urinary system. Evaluating the prognostic outcome at the time of initial diagnosis is essential for patients. Autophagy is known to play a significant role in tumors. Here, we attempted to construct an autophagy-related prognostic risk signature based on the expression profile of autophagy-related genes (ARGs) for predicting the long-term outcome and effect of precise treatments for ccRCC patients. Methods: We obtained the expression profile of ccRCC from the cancer genome atlas (TCGA) database and extract the portion of ARGs. We conducted differentially expressed analysis on ARGs and then performed enrichment analyses to confirm the anomalous autophagy-related biological functions. Then, we performed univariate Cox regression to screen out overall survival (OS)-related ARGs. With these genes, we established an autophagy-related risk signature by least absolute shrinkage and selection operator (LASSO) Cox regression. We validated the reliability of the risk signature with receiver operating characteristic (ROC) analysis, survival analysis, clinic correlation analysis, and Cox regression. Then we analyzed the function of each gene in the signature by single-gene gene set enrichment analysis (GSEA). Finally, we analyzed the correlation between our risk score and expression level of several targets of immunotherapy and targeted therapy. Results: We established a seven-gene prognostic risk signature, according to which we could divide patients into high or low risk groups and predict their outcomes. ROC analysis and survival analysis validated the reliability of the signature. Clinic correlation analysis found that the risk group is significantly correlated with severity of ccRCC. Multivariate Cox regression revealed that the risk score could act as an independent predictor for the prognosis of ccRCC patients. Correlation analysis between risk score and targets of precise treatments showed that our risk signature could predict the effects of precise treatment powerfully. Conclusion: Our study provided a brand new autophagy-related seven-gene prognostic risk signature, which could perform as a prognostic indicator for ccRCC. Meanwhile, our study provides a novel sight to understand the role of autophagy and suggest therapeutic strategies in the category of precise treatment in ccRCC.
Keywords: TCGA; autophagy-related genes; clear cell renal cell carcinoma; effect prediction of precise treatments; least absolute shrinkage and selection operator (LASSO) Cox regression; overall survival; prognostic outcome; prognostic risk signature.
Copyright © 2020 Yang, Han and Li.
Figures




References
LinkOut - more resources
Full Text Sources

