H3K4me1 Distribution Predicts Transcription State and Poising at Promoters
- PMID: 32432110
- PMCID: PMC7214686
- DOI: 10.3389/fcell.2020.00289
H3K4me1 Distribution Predicts Transcription State and Poising at Promoters
Abstract
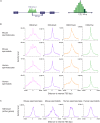
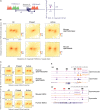
Monomethylation on lysine 4 of histone H3 (H3K4me1) is commonly associated with distal enhancers, but H3K4me1 is also present at promoter regions proximal to transcription start sites. To assess a possible role for H3K4me1 in dictating gene regulatory states at promoters, we examined H3K4me1 peak density around promoters in human and mouse germ cells using an analytic strategy that allowed us to assess relationships between different epigenetic marks on a promoter-by-promoter basis. We found that H3K4me1 exhibits either a bimodal pattern at active promoters, where it flanks H3K4me3, or a unimodal pattern at poised promoters, where it coincides with both H3K4me3 and H3K27me3. This pattern is correlated with gene expression level, but is more strongly linked to a poised chromatin state, defined by the simultaneous presence of H3K4me3 and H3K27me3, than to transcriptional activity. The pattern is especially prominent in germ cells, but is also present in other cell types, including embryonic stem cells and differentiated somatic cells. We propose that H3K4me1 is a key feature of the poised epigenetic state, and suggest possible roles for this mark in epigenetic memory.
Keywords: bivalent; germ cell; histone; pluripotency; poised; promoter; spermatogenesis; stem cell.
Copyright © 2020 Bae and Lesch.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

