CD95 Structure, Aggregation and Cell Signaling
- PMID: 32432115
- PMCID: PMC7214685
- DOI: 10.3389/fcell.2020.00314
CD95 Structure, Aggregation and Cell Signaling
Abstract
CD95 is a pre-ligand-associated transmembrane (TM) receptor. The interaction with its ligand CD95L brings to a next level its aggregation and triggers different signaling pathways, leading to cell motility, differentiation or cell death. This diversity of biological responses associated with a unique receptor devoid of enzymatic property raises the question of whether different ligands exist, or whether the fine-tuned control of CD95 aggregation and conformation, its distribution within certain plasma membrane sub-domains or the pattern of post-translational modifications account for this such broad-range of cell signaling. Herein, we review how the different domains of CD95 and their post-translational modifications or the different forms of CD95L can participate in the receptor aggregation and induction of cell signaling. Understanding how CD95 response goes from cell death to cell proliferation, differentiation and motility is a prerequisite to reveal novel therapeutic options to treat chronic inflammatory disorders and cancers.
Keywords: Fas; aggregation; apoptosis; inflammation; migration; stoichiometry.
Copyright © 2020 Levoin, Jean and Legembre.
Figures



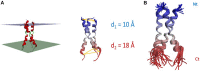


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

