Disease Knowledge Transfer across Neurodegenerative Diseases
- PMID: 32432230
- PMCID: PMC7235145
- DOI: 10.1007/978-3-030-32245-8_95
Disease Knowledge Transfer across Neurodegenerative Diseases
Abstract
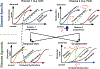
We introduce Disease Knowledge Transfer (DKT), a novel technique for transferring biomarker information between related neurodegenerative diseases. DKT infers robust multimodal biomarker trajectories in rare neurodegenerative diseases even when only limited, unimodal data is available, by transferring information from larger multimodal datasets from common neurodegenerative diseases. DKT is a joint-disease generative model of biomarker progressions, which exploits biomarker relationships that are shared across diseases. Our proposed method allows, for the first time, the estimation of plausible multimodal biomarker trajectories in Posterior Cortical Atrophy (PCA), a rare neurodegenerative disease where only unimodal MRI data is available. For this we train DKT on a combined dataset containing subjects with two distinct diseases and sizes of data available: 1) a larger, multimodal typical AD (tAD) dataset from the TADPOLE Challenge, and 2) a smaller unimodal Posterior Cortical Atrophy (PCA) dataset from the Dementia Research Centre (DRC), for which only a limited number of Magnetic Resonance Imaging (MRI) scans are available. Although validation is challenging due to lack of data in PCA, we validate DKT on synthetic data and two patient datasets (TADPOLE and PCA cohorts), showing it can estimate the ground truth parameters in the simulation and predict unseen biomarkers on the two patient datasets. While we demonstrated DKT on Alzheimer's variants, we note DKT is generalisable to other forms of related neurodegenerative diseases. Source code for DKT is available online: https://github.com/mrazvan22/dkt.
Keywords: Alzheimer’s Disease; Disease Progression Modelling; Manifold Learning; Posterior Cortical Atrophy; Transfer Learning.
Figures

References
-
- Hon M and Khan N, 2017. Towards Alzheimer’s Disease Classification through Transfer Learning. arXiv preprint arXiv:1711.11117.
Grants and funding
LinkOut - more resources
Full Text Sources
