Interaction between microbiota and immunity in health and disease
- PMID: 32433595
- PMCID: PMC7264227
- DOI: 10.1038/s41422-020-0332-7
Interaction between microbiota and immunity in health and disease
Abstract
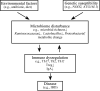
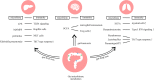
The interplay between the commensal microbiota and the mammalian immune system development and function includes multifold interactions in homeostasis and disease. The microbiome plays critical roles in the training and development of major components of the host's innate and adaptive immune system, while the immune system orchestrates the maintenance of key features of host-microbe symbiosis. In a genetically susceptible host, imbalances in microbiota-immunity interactions under defined environmental contexts are believed to contribute to the pathogenesis of a multitude of immune-mediated disorders. Here, we review features of microbiome-immunity crosstalk and their roles in health and disease, while providing examples of molecular mechanisms orchestrating these interactions in the intestine and extra-intestinal organs. We highlight aspects of the current knowledge, challenges and limitations in achieving causal understanding of host immune-microbiome interactions, as well as their impact on immune-mediated diseases, and discuss how these insights may translate towards future development of microbiome-targeted therapeutic interventions.
Conflict of interest statement
E.E. is a salaried scientific consultant for DayTwo and BiomX. D.Z. and T.L. have nothing to declare.
Figures

References
-
- Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. - PubMed
-
- Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603–616. - PubMed
-
- Lynch JB, Hsiao EY. Microbiomes as sources of emergent host phenotypes. Science. 2019;365:1405–1409. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

