TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7-9
- PMID: 32433612
- PMCID: PMC7610944
- DOI: 10.1038/s41586-020-2282-0
TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7-9
Abstract
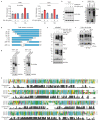
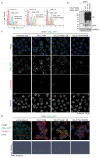
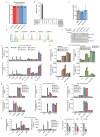
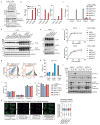
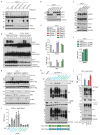
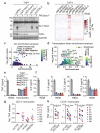
Toll-like receptors (TLRs) have a crucial role in the recognition of pathogens and initiation of immune responses1-3. Here we show that a previously uncharacterized protein encoded by CXorf21-a gene that is associated with systemic lupus erythematosus4,5-interacts with the endolysosomal transporter SLC15A4, an essential but poorly understood component of the endolysosomal TLR machinery also linked to autoimmune disease4,6-9. Loss of this type-I-interferon-inducible protein, which we refer to as 'TLR adaptor interacting with SLC15A4 on the lysosome' (TASL), abrogated responses to endolysosomal TLR agonists in both primary and transformed human immune cells. Deletion of SLC15A4 or TASL specifically impaired the activation of the IRF pathway without affecting NF-κB and MAPK signalling, which indicates that ligand recognition and TLR engagement in the endolysosome occurred normally. Extensive mutagenesis of TASL demonstrated that its localization and function relies on the interaction with SLC15A4. TASL contains a conserved pLxIS motif (in which p denotes a hydrophilic residue and x denotes any residue) that mediates the recruitment and activation of IRF5. This finding shows that TASL is an innate immune adaptor for TLR7, TLR8 and TLR9 signalling, revealing a clear mechanistic analogy with the IRF3 adaptors STING, MAVS and TRIF10,11. The identification of TASL as the component that links endolysosomal TLRs to the IRF5 transcription factor via SLC15A4 provides a mechanistic explanation for the involvement of these proteins in systemic lupus erythematosus12-14.
Conflict of interest statement
JE.L., J.L. J.K., M.L.M. and C.W. are employed in the Immunology & Respiratory department, Drug Concept Discovery group of Boehringer-Ingelheim. The work in the Superti-Furga laboratory is supported by Boehringer-Ingelheim (Grant Agreement 238114).
Figures








Comment in
-
Fourth defence molecule completes antiviral line-up.Nature. 2020 May;581(7808):266-267. doi: 10.1038/d41586-020-01336-2. Nature. 2020. PMID: 32405039 No abstract available.
-
The (Orf)ull truth about IRF5 and type I interferons in SLE.Nat Rev Rheumatol. 2020 Oct;16(10):543-544. doi: 10.1038/s41584-020-0472-7. Nat Rev Rheumatol. 2020. PMID: 32709997 No abstract available.
-
The role of TASL in the pathogenesis of SLE: X marks the spot.Ann Rheum Dis. 2021 Jan;80(1):6-7. doi: 10.1136/annrheumdis-2020-218643. Epub 2020 Sep 25. Ann Rheum Dis. 2021. PMID: 32978239 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

