Cortical Observation by Synchronous Multifocal Optical Sampling Reveals Widespread Population Encoding of Actions
- PMID: 32433908
- PMCID: PMC7687350
- DOI: 10.1016/j.neuron.2020.04.023
Cortical Observation by Synchronous Multifocal Optical Sampling Reveals Widespread Population Encoding of Actions
Abstract
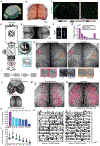
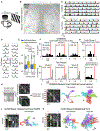
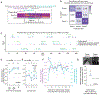
To advance the measurement of distributed neuronal population representations of targeted motor actions on single trials, we developed an optical method (COSMOS) for tracking neural activity in a largely uncharacterized spatiotemporal regime. COSMOS allowed simultaneous recording of neural dynamics at ∼30 Hz from over a thousand near-cellular resolution neuronal sources spread across the entire dorsal neocortex of awake, behaving mice during a three-option lick-to-target task. We identified spatially distributed neuronal population representations spanning the dorsal cortex that precisely encoded ongoing motor actions on single trials. Neuronal correlations measured at video rate using unaveraged, whole-session data had localized spatial structure, whereas trial-averaged data exhibited widespread correlations. Separable modes of neural activity encoded history-guided motor plans, with similar population dynamics in individual areas throughout cortex. These initial experiments illustrate how COSMOS enables investigation of large-scale cortical dynamics and that information about motor actions is widely shared between areas, potentially underlying distributed computations.
Keywords: COSMOS; calcium imaging; cortex; licking; motor planning; multifocal; neural decoding; neural dynamics; population dynamics; synchronous; widefield.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors have made all the designs and protocols for COSMOS freely available for nonprofit use; Stanford University is also submitting a patent application to further facilitate commercial translation.
Figures

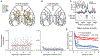


Similar articles
-
Global Representations of Goal-Directed Behavior in Distinct Cell Types of Mouse Neocortex.Neuron. 2017 May 17;94(4):891-907.e6. doi: 10.1016/j.neuron.2017.04.017. Neuron. 2017. PMID: 28521139 Free PMC article.
-
In Vivo Observations of Rapid Scattered Light Changes Associated with Neurophysiological Activity.In: Frostig RD, editor. In Vivo Optical Imaging of Brain Function. 2nd edition. Boca Raton (FL): CRC Press/Taylor & Francis; 2009. Chapter 5. In: Frostig RD, editor. In Vivo Optical Imaging of Brain Function. 2nd edition. Boca Raton (FL): CRC Press/Taylor & Francis; 2009. Chapter 5. PMID: 26844322 Free Books & Documents. Review.
-
Spatial Information Encoding across Multiple Neocortical Regions Depends on an Intact Hippocampus.J Neurosci. 2021 Jan 13;41(2):307-319. doi: 10.1523/JNEUROSCI.1788-20.2020. Epub 2020 Nov 17. J Neurosci. 2021. PMID: 33203745 Free PMC article.
-
Recording human electrocorticographic (ECoG) signals for neuroscientific research and real-time functional cortical mapping.J Vis Exp. 2012 Jun 26;(64):3993. doi: 10.3791/3993. J Vis Exp. 2012. PMID: 22782131 Free PMC article.
-
Comprehensive imaging of cortical networks.Curr Opin Neurobiol. 2015 Jun;32:115-23. doi: 10.1016/j.conb.2015.03.016. Epub 2015 Apr 13. Curr Opin Neurobiol. 2015. PMID: 25880117 Review.
Cited by
-
Simultaneous, cortex-wide and cellular-resolution neuronal population dynamics reveal an unbounded scaling of dimensionality with neuron number.bioRxiv [Preprint]. 2024 Jan 16:2024.01.15.575721. doi: 10.1101/2024.01.15.575721. bioRxiv. 2024. Update in: Neuron. 2024 May 15;112(10):1694-1709.e5. doi: 10.1016/j.neuron.2024.02.011. PMID: 38293036 Free PMC article. Updated. Preprint.
-
The diversity and specificity of functional connectivity across spatial and temporal scales.Neuroimage. 2021 Dec 15;245:118692. doi: 10.1016/j.neuroimage.2021.118692. Epub 2021 Oct 29. Neuroimage. 2021. PMID: 34751153 Free PMC article. Review.
-
Wide-Field Calcium Imaging of Dynamic Cortical Networks during Locomotion.Cereb Cortex. 2022 Jun 7;32(12):2668-2687. doi: 10.1093/cercor/bhab373. Cereb Cortex. 2022. PMID: 34689209 Free PMC article.
-
Mesoscale volumetric light-field (MesoLF) imaging of neuroactivity across cortical areas at 18 Hz.Nat Methods. 2023 Apr;20(4):600-609. doi: 10.1038/s41592-023-01789-z. Epub 2023 Feb 23. Nat Methods. 2023. PMID: 36823333 Free PMC article.
-
The information bottleneck as a principle underlying multi-area cortical representations during decision-making.bioRxiv [Preprint]. 2025 Feb 15:2023.07.12.548742. doi: 10.1101/2023.07.12.548742. bioRxiv. 2025. PMID: 37502862 Free PMC article. Preprint.
References
-
- Akrami A, Kopec CD, Diamond ME, and Brody CD (2018). Posterior parietal cortex represents sensory history and mediates its effects on behaviour. Nature 554, 368–372. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

