AID in Antibody Diversification: There and Back Again
- PMID: 32434680
- PMCID: PMC7183997
- DOI: 10.1016/j.it.2020.04.009
AID in Antibody Diversification: There and Back Again
Erratum in
-
AID in Antibody Diversification: There and Back Again: (Trends in Immunology 41, 586-600; 2020).Trends Immunol. 2021 Jan;42(1):89. doi: 10.1016/j.it.2020.10.011. Epub 2020 Nov 14. Trends Immunol. 2021. PMID: 33203546 Free PMC article. No abstract available.
Abstract
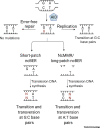
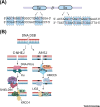
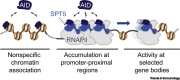
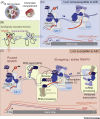
Activation-Induced cytidine Deaminase (AID) initiates affinity maturation and isotype switching by deaminating deoxycytidines within immunoglobulin genes, leading to somatic hypermutation (SHM) and class switch recombination (CSR). AID thus potentiates the humoral response to clear pathogens. Marking the 20th anniversary of the discovery of AID, we review the current understanding of AID function. We discuss AID biochemistry and how error-free forms of DNA repair are co-opted to prioritize mutagenesis over accuracy during antibody diversification. We discuss the regulation of DNA double-strand break (DSB) repair pathways during CSR. We describe genomic targeting of AID as a multilayered process involving chromatin architecture, cis- and trans-acting factors, and determining mutagenesis - distinct from AID occupancy at loci that are spared from mutation.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
References
-
- Schatz D.G. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. - PubMed
-
- Peters A., Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. - PubMed
-
- Rogozin I.B., Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 2004;172:3382–3384. - PubMed
-
- Muramatsu M. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. - PubMed
-
- Revy P. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

