Pseudouridine-mediated stop codon readthrough in S. cerevisiae is sequence context-independent
- PMID: 32434780
- PMCID: PMC7430670
- DOI: 10.1261/rna.076042.120
Pseudouridine-mediated stop codon readthrough in S. cerevisiae is sequence context-independent
Abstract
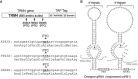
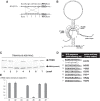
We have previously shown that when the uridine of a stop codon (UAA, UAG, or UGA) is pseudouridylated, the ribosome reads through the modified stop codon. However, it is not clear as to whether or not the pseudouridine (Ψ)-mediated readthrough is dependent on the sequence context of mRNA. Here, we use several different approaches and the yeast system to address this question. We show that when a stop codon (premature termination codon, PTC) is introduced into the coding region of a reporter mRNA at several different positions (with different sequence contexts) and pseudouridylated, we detect similar levels of readthrough. Using mutational and selection/screen analyses, we also show that the upstream sequence (relative to PTC) as well as the nucleotides surrounding the PTC (upstream and downstream) play a minimal role (if at all) in Ψ-mediated ribosome readthrough. Interestingly, we detect no suppression of NMD (nonsense-mediated mRNA decay) by targeted PTC pseudouridylation in the yeast system. Our results indicate that Ψ-mediated nonsense suppression occurs at the translational level, and that the suppression is sequence context-independent, unlike some previously characterized rare stop codon readthrough events.
Keywords: PTC; nonsense suppression; pseudouridylation; sequence context; stop codon.
© 2020 Adachi and Yu; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
