Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification
- PMID: 32435053
- PMCID: PMC7239890
- DOI: 10.1038/s41392-020-0186-y
Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification
Abstract
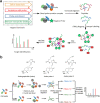
Natural products are an important source of new drugs for the treatment of various diseases. However, developing natural product-based new medicines through random moiety modification is a lengthy and costly process, due in part to the difficulties associated with comprehensively understanding the mechanism of action and the side effects. Identifying the protein targets of natural products is an effective strategy, but most medicines interact with multiple protein targets, which complicate this process. In recent years, an increasing number of researchers have begun to screen the target proteins of natural products with chemical proteomics approaches, which can provide a more comprehensive array of the protein targets of active small molecules in an unbiased manner. Typically, chemical proteomics experiments for target identification consist of two key steps: (1) chemical probe design and synthesis and (2) target fishing and identification. In recent decades, five different types of chemical proteomic probes and their respective target fishing methods have been developed to screen targets of molecules with different structures, and a variety of protein identification approaches have been invented. Presently, we will classify these chemical proteomics approaches, the application scopes and characteristics of the different types of chemical probes, the different protein identification methods, and the advantages and disadvantages of these strategies.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–477. - PubMed
-
- Rodrigues T, Reker D, Schneider P, Schneider G. Counting on natural products for drug design. Nat. Chem. 2016;8:531. - PubMed
-
- Gouyette A. Synthesis of deuterium-labelled elliptinium and its use in metabolic studies. Biomed. Environ. Mass Spectrom. 1988;15:243–247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

