Differential Expression of IgM and IgD Discriminates Two Subpopulations of Human Circulating IgM+IgD+CD27+ B Cells That Differ Phenotypically, Functionally, and Genetically
- PMID: 32435242
- PMCID: PMC7219516
- DOI: 10.3389/fimmu.2020.00736
Differential Expression of IgM and IgD Discriminates Two Subpopulations of Human Circulating IgM+IgD+CD27+ B Cells That Differ Phenotypically, Functionally, and Genetically
Abstract
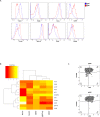
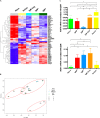
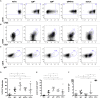
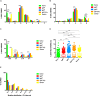
The origin and function of blood IgM+IgD+CD27+ B cells is controversial, and they are considered a heterogeneous population. Previous staining of circulating B cells of healthy donors with rotavirus fluorescent virus-like particles allowed us to differentiate two subsets of IgM+IgD+CD27+: IgMhi and IgMlo B cells. Here, we confirmed this finding and compared the phenotype, transcriptome, in vitro function, and Ig gene repertoire of these two subsets. Eleven markers phenotypically discriminated both subsets (CD1c, CD69, IL21R, CD27, MTG, CD45RB, CD5, CD184, CD23, BAFFR, and CD38) with the IgMhi phenotypically resembling previously reported marginal zone B cells and the IgMlo resembling both naïve and memory B cells. Transcriptomic analysis showed that both subpopulations clustered close to germinal center-experienced IgM only B cells with a Principal Component Analysis, but differed in expression of 78 genes. Moreover, IgMhi B cells expressed genes characteristic of previously reported marginal zone B cells. After stimulation with CpG and cytokines, significantly (p < 0.05) higher frequencies (62.5%) of IgMhi B cells proliferated, compared with IgMlo B cells (35.37%), and differentiated to antibody secreting cells (14.22% for IgMhi and 7.19% for IgMlo). IgMhi B cells had significantly (p < 0.0007) higher frequencies of mutations in IGHV and IGKV regions, IgMlo B cells had higher usage of IGHJ6 genes (p < 0.0001), and both subsets differed in their HCDR3 properties. IgMhi B cells shared most of their shared IGH clonotypes with IgM only memory B cells, and IgMlo B cells with IgMhi B cells. These results support the notion that differential expression of IgM and IgD discriminates two subpopulations of human circulating IgM+IgD+CD27+ B cells, with the IgMhi B cells having similarities with previously described marginal zone B cells that passed through germinal centers, and the IgMlo B cells being the least differentiated amongst the IgM+CD27+ subsets.
Keywords: Ig gene repertoire; blood; cell proliferation; cell surface molecules; gene expression; human; marginal zone B cells; memory B cells.
Copyright © 2020 Bautista, Vásquez, Ayala-Ramírez, Téllez-Sosa, Godoy-Lozano, Martínez-Barnetche, Franco and Angel.
Figures


References
-
- Tian C, Luskin GK, Dischert KM, Higginbotham JN, Shepherd BE, Crowe JE. Immunodominance of the VH1-46 antibody gene segment in the primary repertoire of human rotavirus-specific B cells is reduced in the memory compartment through somatic mutation of nondominant clones. J Immunol. (2008) 180:3279–88. 10.4049/jimmunol.180.5.3279 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

