Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data
- PMID: 32438586
- PMCID: PMC7290575
- DOI: 10.3390/v12050560
Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data
Abstract
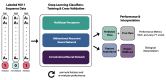
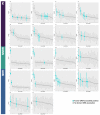
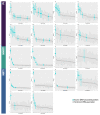
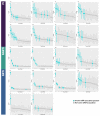
The fast replication rate and lack of repair mechanisms of human immunodeficiency virus (HIV) contribute to its high mutation frequency, with some mutations resulting in the evolution of resistance to antiretroviral therapies (ART). As such, studying HIV drug resistance allows for real-time evaluation of evolutionary mechanisms. Characterizing the biological process of drug resistance is also critically important for sustained effectiveness of ART. Investigating the link between "black box" deep learning methods applied to this problem and evolutionary principles governing drug resistance has been overlooked to date. Here, we utilized publicly available HIV-1 sequence data and drug resistance assay results for 18 ART drugs to evaluate the performance of three architectures (multilayer perceptron, bidirectional recurrent neural network, and convolutional neural network) for drug resistance prediction, jointly with biological analysis. We identified convolutional neural networks as the best performing architecture and displayed a correspondence between the importance of biologically relevant features in the classifier and overall performance. Our results suggest that the high classification performance of deep learning models is indeed dependent on drug resistance mutations (DRMs). These models heavily weighted several features that are not known DRM locations, indicating the utility of model interpretability to address causal relationships in viral genotype-phenotype data.
Keywords: HIV; HIV drug resistance; antiretroviral therapy; deep learning; machine learning; neural networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Using machine learning and big data to explore the drug resistance landscape in HIV.PLoS Comput Biol. 2021 Aug 26;17(8):e1008873. doi: 10.1371/journal.pcbi.1008873. eCollection 2021 Aug. PLoS Comput Biol. 2021. PMID: 34437532 Free PMC article.
-
Changes from 2000 to 2009 in the Prevalence of HIV-1 Containing Drug Resistance-Associated Mutations from Antiretroviral Therapy-Naive, HIV-1-Infected Patients in the United States.AIDS Res Hum Retroviruses. 2018 Aug;34(8):672-679. doi: 10.1089/AID.2017.0295. Epub 2018 Jun 5. AIDS Res Hum Retroviruses. 2018. PMID: 29732898 Free PMC article.
-
Natural polymorphisms in HIV-1 CRF01_AE strain and profile of acquired drug resistance mutations in a long-term combination treatment cohort in northeastern China.BMC Infect Dis. 2020 Feb 26;20(1):178. doi: 10.1186/s12879-020-4808-3. BMC Infect Dis. 2020. PMID: 32102660 Free PMC article.
-
Current perspectives on HIV-1 antiretroviral drug resistance.Viruses. 2014 Oct 24;6(10):4095-139. doi: 10.3390/v6104095. Viruses. 2014. PMID: 25341668 Free PMC article. Review.
-
Impact of human immunodeficiency virus type-1 sequence diversity on antiretroviral therapy outcomes.Viruses. 2014 Oct 20;6(10):3855-72. doi: 10.3390/v6103855. Viruses. 2014. PMID: 25333465 Free PMC article. Review.
Cited by
-
Role of Artificial Intelligence and Personalized Medicine in Enhancing HIV Management and Treatment Outcomes.Life (Basel). 2025 May 6;15(5):745. doi: 10.3390/life15050745. Life (Basel). 2025. PMID: 40430173 Free PMC article. Review.
-
Single-Cell Techniques and Deep Learning in Predicting Drug Response.Trends Pharmacol Sci. 2020 Dec;41(12):1050-1065. doi: 10.1016/j.tips.2020.10.004. Epub 2020 Nov 2. Trends Pharmacol Sci. 2020. PMID: 33153777 Free PMC article. Review.
-
Artificial Intelligence, Machine Learning, and Big Data for Ebola Virus Drug Discovery.Pharmaceuticals (Basel). 2023 Feb 21;16(3):332. doi: 10.3390/ph16030332. Pharmaceuticals (Basel). 2023. PMID: 36986432 Free PMC article.
-
Web Service for HIV Drug Resistance Prediction Based on Analysis of Amino Acid Substitutions in Main Drug Targets.Viruses. 2023 Nov 11;15(11):2245. doi: 10.3390/v15112245. Viruses. 2023. PMID: 38005921 Free PMC article.
-
Assessment of a Computational Approach to Predict Drug Resistance Mutations for HIV, HBV and SARS-CoV-2.Molecules. 2022 Aug 24;27(17):5413. doi: 10.3390/molecules27175413. Molecules. 2022. PMID: 36080181 Free PMC article.
References
-
- Centers for Disease Control and Prevention . HIV Surveillance Report. Volume 30 Centers for Disease Control and Prevention; Atlanta, GA, USA: 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

