Role of the trigger loop in translesion RNA synthesis by bacterial RNA polymerase
- PMID: 32439804
- PMCID: PMC7363142
- DOI: 10.1074/jbc.RA119.011844
Role of the trigger loop in translesion RNA synthesis by bacterial RNA polymerase
Abstract
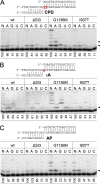
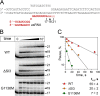
DNA lesions can severely compromise transcription and block RNA synthesis by RNA polymerase (RNAP), leading to subsequent recruitment of DNA repair factors to the stalled transcription complex. Recent structural studies have uncovered molecular interactions of several DNA lesions within the transcription elongation complex. However, little is known about the role of key elements of the RNAP active site in translesion transcription. Here, using recombinantly expressed proteins, in vitro transcription, kinetic analyses, and in vivo cell viability assays, we report that point amino acid substitutions in the trigger loop, a flexible element of the active site involved in nucleotide addition, can stimulate translesion RNA synthesis by Escherichia coli RNAP without altering the fidelity of nucleotide incorporation. We show that these substitutions also decrease transcriptional pausing and strongly affect the nucleotide addition cycle of RNAP by increasing the rate of nucleotide addition but also decreasing the rate of translocation. The secondary channel factors DksA and GreA modulated translesion transcription by RNAP, depending on changes in the trigger loop structure. We observed that although the mutant RNAPs stimulate translesion synthesis, their expression is toxic in vivo, especially under stress conditions. We conclude that the efficiency of translesion transcription can be significantly modulated by mutations affecting the conformational dynamics of the active site of RNAP, with potential effects on cellular stress responses and survival.
Keywords: DNA damage; DNA repair; DksA; GreA; RNA; RNA polymerase; RNA polymerase (RNAP); nucleotide excision repair (NER); secondary channel factor; transcription; transcription-coupled repair; translesion transcription; trigger loop.
© 2020 Agapov et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

