Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction
- PMID: 32439900
- PMCID: PMC7242331
- DOI: 10.1038/s41467-020-15706-x
Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction
Abstract
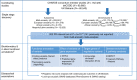
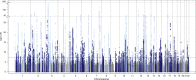
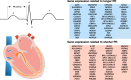
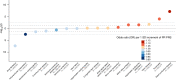
The electrocardiographic PR interval reflects atrioventricular conduction, and is associated with conduction abnormalities, pacemaker implantation, atrial fibrillation (AF), and cardiovascular mortality. Here we report a multi-ancestry (N = 293,051) genome-wide association meta-analysis for the PR interval, discovering 202 loci of which 141 have not previously been reported. Variants at identified loci increase the percentage of heritability explained, from 33.5% to 62.6%. We observe enrichment for cardiac muscle developmental/contractile and cytoskeletal genes, highlighting key regulation processes for atrioventricular conduction. Additionally, 8 loci not previously reported harbor genes underlying inherited arrhythmic syndromes and/or cardiomyopathies suggesting a role for these genes in cardiovascular pathology in the general population. We show that polygenic predisposition to PR interval duration is an endophenotype for cardiovascular disease, including distal conduction disease, AF, and atrioventricular pre-excitation. These findings advance our understanding of the polygenic basis of cardiac conduction, and the genetic relationship between PR interval duration and cardiovascular disease.
Conflict of interest statement
I.N. became a full-time employee of Gilead Sciences Ltd following submission of the manuscript. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, and Boehringer Ingelheim, and has consulted for Bristol Myers Squibb/Pfizer and Bayer AG. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases. P.T.E. has also served on advisory boards or consulted for Bayer AG, Quest Diagnostics, and Novartis. M.J.C. is Chief Scientist for Genomics England, a UK Government company. B.M.P. serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. V.S. has participated in a conference trip sponsored by Novo Nordisk and received a modest honorarium for participating in an advisory board meeting. K.S., H.H., P.S., G.S., G.T., R.B.T., U.T., D.O.A., D.F.G. are employed by deCODE genetics/Amgen Inc. E.I. is employed by GlaxoSmithKline. A.M. is employed by Genentech Inc.
Figures
References
Publication types
MeSH terms
Grants and funding
- R01 HL139731/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- MR/N025083/1/MRC_/Medical Research Council/United Kingdom
- U01 HL120393/HL/NHLBI NIH HHS/United States
- K01 HL140187/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- P30 ES007033/ES/NIEHS NIH HHS/United States
- K24 HL148521/HL/NHLBI NIH HHS/United States
- PG/20/18/35058/BHF_/British Heart Foundation/United Kingdom
- U01 HL137162/HL/NHLBI NIH HHS/United States
- PG/17/59/33139/BHF_/British Heart Foundation/United Kingdom
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- G9521010/MRC_/Medical Research Council/United Kingdom
- R01 HL092577/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

