Mapping the epigenetic modifications of DNA and RNA
- PMID: 32440736
- PMCID: PMC7647981
- DOI: 10.1007/s13238-020-00733-7
Mapping the epigenetic modifications of DNA and RNA
Abstract
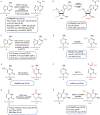
Over 17 and 160 types of chemical modifications have been identified in DNA and RNA, respectively. The interest in understanding the various biological functions of DNA and RNA modifications has lead to the cutting-edged fields of epigenomics and epitranscriptomics. Developing chemical and biological tools to detect specific modifications in the genome or transcriptome has greatly facilitated their study. Here, we review the recent technological advances in this rapidly evolving field. We focus on high-throughput detection methods and biological findings for these modifications, and discuss questions to be addressed as well. We also summarize third-generation sequencing methods, which enable long-read and single-molecule sequencing of DNA and RNA modification.
Keywords: DNA methylation; DNA modification; RNA modification; epigenetics; epitranscriptomics; long read sequencing.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

