Scalable, two-stage, autoinduction of recombinant protein expression in E. coli utilizing phosphate depletion
- PMID: 32441815
- PMCID: PMC9589519
- DOI: 10.1002/bit.27440
Scalable, two-stage, autoinduction of recombinant protein expression in E. coli utilizing phosphate depletion
Abstract
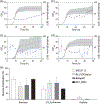
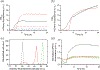
We report the scalable production of recombinant proteins in Escherichia coli, reliant on tightly controlled autoinduction, triggered by phosphate depletion in the stationary phase. The method, reliant on engineered strains and plasmids, enables improved protein expression across scales. Expression levels using this approach have reached as high as 55% of the total cellular protein. The initial use of the method in instrumented fed-batch fermentations enables cell densities of ∼30 gCDW/L and protein titers up to 8.1 ± 0.7 g/L (∼270 mg/gCDW). The process has also been adapted to an optimized autoinduction media, enabling routine batch production at culture volumes of 20 μl (384-well plates), 100 μl (96-well plates), 20 ml, and 100 ml. In batch cultures, cell densities routinely reach ∼5-7 gCDW/L, offering protein titers above 2 g/L. The methodology has been validated with a set of diverse heterologous proteins and is of general use for the facile optimization of routine protein expression from high throughput screens to fed-batch fermentation.
Keywords: autoinduction; phosphate depletion; protein expression; stationary phase.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTERESTS
Michael D. Lynch and Zhixia Ye have a financial interest in DMC Biotechnologies Inc. Romel Menacho-Melgar, Zhixia Ye, and Michael D. Lynch have filed patent applications on strains and methods discussed in this manuscript. Other authors declare that there are no conflict of interests.
Figures



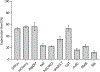
References
-
- An JH, & Kim YS (1998). A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii—Cloning, sequencing, and expression of the enzymes in Escherichia coli. European Journal of Biochemistry, 257(2), 395–402. - PubMed
-
- Anilionyte O, Liang H, Ma X, Yang L, & Zhou K (2018). Short, auto-inducible promoters for well-controlled protein expression in Escherichia coli. Applied Microbiology and Biotechnology, 102(16), 7007–7015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- YIP #N00014-16-1-2558/Office of Naval Research/International
- EAGER: #1445726/National Science Foundation/International
- HR0011‐14‐C‐0075/Defense Advanced Research Projects Agency/International
- #EE0007563/Department of Energy EERE/International
- 2018-BIG-6503/North Carolina Biotechnology Center/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

