Hepatitis B Virus-X Downregulates Expression of Selenium Binding Protein 1
- PMID: 32443734
- PMCID: PMC7291177
- DOI: 10.3390/v12050565
Hepatitis B Virus-X Downregulates Expression of Selenium Binding Protein 1
Abstract
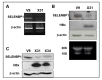
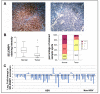
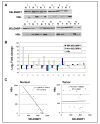
Selenium binding protein 1 (SELENBP1) has been known to be reduced in various types cancer, and epigenetic change is shown to be likely to account for the reduction of SELNEBP1 expression. With cDNA microarray comparative analysis, we found that SELENBP1 is markedly decreased in hepatitis B virus-X (HBx)-expressing cells. To clarify the effect of HBx on SELENBP1 expression, we compared the expression levels of SELENBP1 mRNA and protein by semi-quantitative RT-PCR, Northern blot, and Western blot. As expected, SELENBP1 expression was shown to be reduced in cells expressing HBx, and reporter gene analysis showed that the SELENBP1 promoter is repressed by HBx. In addition, the stepwise deletion of 5' flanking promoter sequences resulted in a gradual decrease in basal promoter activity and inhibition of SELENBP1 expression by HBx. Moreover, immunohistochemistry on tissue microarrays containing 60 pairs of human liver tissue showed decreased intensity of SELENBP1 in tumor tissues as compared with their matched non-tumor liver tissues. Taken together, our findings suggest that inhibition of SELENBP1 expression by HBx might act as one of the causes in the development of hepatocellular carcinoma caused by HBV infection.
Keywords: X protein; hepatitis B virus; hepatocellular carcinoma; selenium binding protein 1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Torrealba J.R., Colburn M., Golner S., Chang Z., Scheunemann T., Fechner J.H., Roenneburg D., Hu H., Alam T., Kim H.T. Selenium-binding protein1 in smooth muscle cells is downregulated in a Rhesus monkey model of chronic allograft nephropathy. Am. J. Transplant. 2005;5:58–67. doi: 10.1111/j.1600-6143.2004.00651.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

