MicroRNA-1253 Regulation of WASF2 (WAVE2) and its Relevance to Racial Health Disparities
- PMID: 32443852
- PMCID: PMC7288301
- DOI: 10.3390/genes11050572
MicroRNA-1253 Regulation of WASF2 (WAVE2) and its Relevance to Racial Health Disparities
Abstract
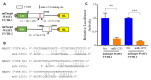
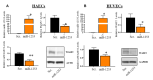
The prevalence of hypertension among African Americans (AAs) in the US is among the highest of any demographic and affects over two-thirds of AA women. Previous data from our laboratory suggest substantial differential gene expression (DGE) of mRNAs and microRNAs (miRNAs) exists within peripheral blood mononuclear cells (PBMCs) isolated from AA and white women with or without hypertension. We hypothesized that DGE by race may contribute to racial differences in hypertension. In a reanalysis of our previous dataset, we found that the Wiskott-Aldrich syndrome protein Verprolin-homologous protein 2 (WASF2 (also known as WAVE2)) is differentially expressed in AA women with hypertension, along with several other members of the actin cytoskeleton signaling pathway that plays a role in cell shape and branching of actin filaments. We performed an in silico miRNA target prediction analysis that suggested miRNA miR-1253 regulates WASF2. Transfection of miR-1253 mimics into human umbilical vein endothelial cells (HUVECs) and human aortic endothelial cells (HAECs) significantly repressed WASF2 mRNA and protein levels (p < 0.05), and a luciferase reporter assay confirmed that miR-1253 regulates the WASF2 3' UTR (p < 0.01). miR-1253 overexpression in HUVECs significantly increased HUVEC lamellipodia formation (p < 0.01), suggesting the miR-1253-WASF2 interaction may play a role in cell shape and actin cytoskeleton function. Together, we have identified novel roles for miR-1253 and WASF2 in a hypertension-related disparities context. This may ultimately lead to the discovery of additional actin-related genes which are important in the vascular-related complications of hypertension and influence the disproportionate susceptibility to hypertension among AAs in general and AA women in particular.
Keywords: African American; actin cytoskeletal regulators; differential gene expression; endothelial cell; health disparities; hypertension; microRNA; race; women.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Benjamin E.J., Muntner A., Alonso M.S., Bittencourt C.W., Callaway A.P., Carson A.M., Chamberlain A.R., Chang S., Cheng S.R., Das F.N., et al. Virani, Epidemiology American Heart Association Council on, Committee Prevention Statistics, and Subcommittee Stroke Statistics. Heart Disease and Stroke Statistics-2019 Update: A Report from the American Heart Association. Circulation. 2019;139:E56–E528. doi: 10.1161/CIR.0000000000000659. - DOI - PubMed
-
- Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.-C., Freiman A., Sams A.J., Hebert S., et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. - DOI - PubMed
-
- Wang B.-D., Ceniccola K., Yang Q., Andrawis R., Patel V., Ji Y., Rhim J., Olender J., Popratiloff A., Latham P., et al. Identification and Functional Validation of Reciprocal microRNA-mRNA Pairings in African American Prostate Cancer Disparities. Clin. Cancer Res. 2015;21:4970–4984. doi: 10.1158/1078-0432.CCR-14-1566. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

