Methanol-Essential Growth of Corynebacterium glutamicum: Adaptive Laboratory Evolution Overcomes Limitation due to Methanethiol Assimilation Pathway
- PMID: 32443885
- PMCID: PMC7279501
- DOI: 10.3390/ijms21103617
Methanol-Essential Growth of Corynebacterium glutamicum: Adaptive Laboratory Evolution Overcomes Limitation due to Methanethiol Assimilation Pathway
Abstract
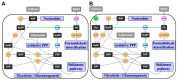
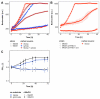
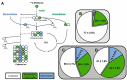
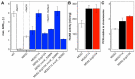
Methanol is a sustainable substrate for biotechnology. In addition to natural methylotrophs, metabolic engineering has gained attention for transfer of methylotrophy. Here, we engineered Corynebacterium glutamicum for methanol-dependent growth with a sugar co-substrate. Heterologous expression of genes for methanol dehydrogenase from Bacillus methanolicus and of ribulose monophosphate pathway genes for hexulose phosphate synthase and isomerase from Bacillus subtilis enabled methanol-dependent growth of mutants carrying one of two independent metabolic cut-offs, i.e., either lacking ribose-5-phosphate isomerase or ribulose-5-phosphate epimerase. Whole genome sequencing of strains selected by adaptive laboratory evolution (ALE) for faster methanol-dependent growth was performed. Subsequently, three mutations were identified that caused improved methanol-dependent growth by (1) increased plasmid copy numbers, (2) enhanced riboflavin supply and (3) reduced formation of the methionine-analogue O-methyl-homoserine in the methanethiol pathway. Our findings serve as a foundation for the engineering of C. glutamicum to unleash the full potential of methanol as a carbon source in biotechnological processes.
Keywords: adaptive laboratory evolution; isotopic labeling; metabolic engineering; methanol; ribulose monophosphate pathway; synthetic methylotrophy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

