Identification of Potential Oral Microbial Biomarkers for the Diagnosis of Periodontitis
- PMID: 32443919
- PMCID: PMC7290295
- DOI: 10.3390/jcm9051549
Identification of Potential Oral Microbial Biomarkers for the Diagnosis of Periodontitis
Abstract
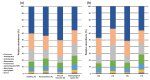
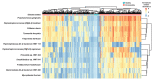
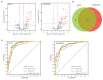
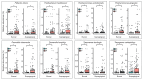
Periodontitis is a chronic and multifactorial inflammatory disease that can lead to tooth loss. At present, the diagnosis for periodontitis is primarily based on clinical examination and radiographic parameters. Detecting the periodontal pathogens at the subgingival plaque requires skilled professionals to collect samples. Periodontal pathogens are also detected on various mucous membranes in patients with periodontitis. In this study, we characterized the oral microbiome profiles from buccal mucosa and supragingival space in a total of 272 healthy subjects as a control group, and periodontitis patients as a disease group. We identified 13 phyla, 193 genera, and 527 species and determined periodontitis-associated taxa. Porphyromonas gingivalis, Tannerella forsythia, Treponema denticolar, Filifactor alocis, Porphyromonas endodontalis, Fretibacterium fastiosum and Peptostreptococcus species were significantly increased in both the buccal mucosa and the supragingival space in periodontitis patients. The identified eight periodontitis-associated bacterial species were clinically validated in an independent cohort. We generated the prediction model based on the oral microbiome profiles using five machine learning algorithms, and validated its capability in predicting the status of patients with periodontitis. The results showed that the oral microbiome profiles from buccal mucosa and supragingival space can represent the microbial composition of subgingival plaque and further be utilized to identify potential microbial biomarkers for the diagnosis of periodontitis. Besides, bacterial community interaction network analysis found distinct patterns associated with dysbiosis in periodontitis. In summary, we have identified oral bacterial species from buccal and supragingival sites which can predict subgingival bacterial composition and can be used for early diagnosis of periodontitis. Therefore, our study provides an important basis for developing easy and noninvasive methods to diagnose and monitor periodontitis.
Keywords: Bioinformatics; Biomarkers; Microbiome; Oral bacteria; Periodontal disease(s)/periodontitis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

