Support Vector Machine for Lung Adenocarcinoma Staging Through Variant Pathways
- PMID: 32444360
- PMCID: PMC7341118
- DOI: 10.1534/g3.120.401207
Support Vector Machine for Lung Adenocarcinoma Staging Through Variant Pathways
Abstract
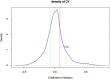
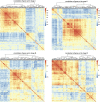
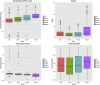
Lung adenocarcinoma (LUAD) is one of the most common malignant tumors. How to effectively diagnose LUAD at an early stage and make an accurate judgement of the occurrence and progression of LUAD are still the focus of current research. Support vector machine (SVM) is one of the most effective methods for diagnosing LUAD of different stages. The study aimed to explore the dynamic change of differentially expressed genes (DEGs) in different stages of LUAD, and to assess the risk of LUAD through DEGs enriched pathways and establish a diagnostic model based on SVM method. Based on TMN stages and gene expression profiles of 517 samples in TCGA-LUAD database, coefficient of variation (CV) combined with one-way analysis of variance (ANOVA) were used to screen out feature genes in different TMN stages after data standardization. Unsupervised clustering analysis was conducted on samples and feature genes. The feature genes were analyzed by Pearson correlation coefficient to construct a co-expression network. Fisher exact test was conducted to verify the most enriched pathways, and the variation of each pathway in different stages was analyzed. SVM networks were trained and ROC curves were drawn based on the predicted results so as to evaluate the predictive effectiveness of the SVM model. Unsupervised hierarchical clustering analysis results showed that almost all the samples in stage III/IV were clustered together, while samples in stage I/II were clustered together. The correlation of feature genes in different stages was different. In addition, with the increase of malignant degree of lung cancer, the average shortest path of the network gradually increased, while the closeness centrality gradually decreased. Finally, four feature pathways that could distinguish different stages of LUAD were obtained and the ability was tested by the SVM model with an accuracy of 91%. Functional level differences were quantified based on the expression of feature genes in lung cancer patients of different stages, so as to help the diagnosis and prediction of lung cancer. The accuracy of our model in differentiating between stage I/II and stage III/IV could reach 91%.
Keywords: co-expression; diagnostic model; functional pathway; lung adenocarcinoma.
Copyright © 2020 Di et al.
Figures







References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
