IRF5 guides monocytes toward an inflammatory CD11c+ macrophage phenotype and promotes intestinal inflammation
- PMID: 32444476
- PMCID: PMC7611075
- DOI: 10.1126/sciimmunol.aax6085
IRF5 guides monocytes toward an inflammatory CD11c+ macrophage phenotype and promotes intestinal inflammation
Abstract
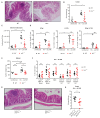
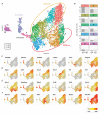
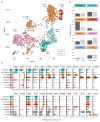
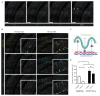
Mononuclear phagocytes (MNPs) are vital for maintaining intestinal homeostasis but, in response to acute microbial stimulation, can also trigger immunopathology, accelerating recruitment of Ly6Chi monocytes to the gut. The regulators that control monocyte tissue adaptation in the gut remain poorly understood. Interferon regulatory factor 5 (IRF5) is a transcription factor previously shown to play a key role in maintaining the inflammatory phenotype of macrophages. Here, we investigate the impact of IRF5 on the MNP system and physiology of the gut at homeostasis and during inflammation. We demonstrate that IRF5 deficiency has a limited impact on colon physiology at steady state but ameliorates immunopathology during Helicobacter hepaticus-induced colitis. Inhibition of IRF5 activity in MNPs phenocopies global IRF5 deficiency. Using a combination of bone marrow chimera and single-cell RNA-sequencing approaches, we examined the intrinsic role of IRF5 in controlling colonic MNP development. We demonstrate that IRF5 promotes differentiation of Ly6Chi monocytes into CD11c+ macrophages and controls the production of antimicrobial and inflammatory mediators by these cells. Thus, we identify IRF5 as a key transcriptional regulator of the colonic MNP system during intestinal inflammation.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
FP received research funding or consultancy fees from GSK, Genentech, Roche and UCB. Other authors declare that they have no competing interests.
Figures




References
-
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298. - PubMed
-
- Bryant RV, Brain O, Travis SPL. Conventional drug therapy for inflammatory bowel disease. Scandinavian Journal of Gastroenterology. 2015;50:90–112. - PubMed
-
- Bravatà I, Fiorino G, Allocca M, Repici A, Danese S. New targeted therapies such as anti-adhesion molecules, anti-IL-12/23 and anti-Janus kinases are looking toward a more effective treatment of inflammatory bowel disease. Scandinavian Journal of Gastroenterology. 2015;50:113–120. - PubMed
-
- Meuret G, Bitzi A, Hammer B. Macrophage turnover in Crohn’s disease and ulcerative colitis. Gastroenterology. 1978;74:501–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

