Chromatin as a key consumer in the metabolite economy
- PMID: 32444835
- PMCID: PMC7258299
- DOI: 10.1038/s41589-020-0517-x
Chromatin as a key consumer in the metabolite economy
Abstract
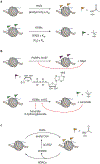
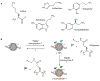
In eukaryotes, chromatin remodeling and post-translational modifications (PTMs) shape the local chromatin landscape to establish permissive and repressive regions within the genome, orchestrating transcription, replication, and DNA repair in concert with other epigenetic mechanisms. Though cellular nutrient signaling encompasses a huge number of pathways, recent attention has turned to the hypothesis that the metabolic state of the cell is communicated to the genome through the type and concentration of metabolites in the nucleus that are cofactors for chromatin-modifying enzymes. Importantly, both epigenetic and metabolic dysregulation are hallmarks of a range of diseases, and this metabolism-chromatin axis may yield a well of new therapeutic targets. In this Perspective, we highlight emerging themes in the inter-regulation of the genome and metabolism via chromatin, including nonenzymatic histone modifications arising from chemically reactive metabolites, the expansion of PTM diversity from cofactor-promiscuous chromatin-modifying enzymes, and evidence for the existence and importance of subnucleocytoplasmic metabolite pools.
Conflict of interest statement
Competing Financial Interests Statement
The authors declare no competing interests.
Figures




References
-
- Kornberg RD Structure of Chromatin. Annu. Rev. Biochem 46, 931–954 (1977). - PubMed
-
- Kouzarides T Chromatin Modifications and Their Function. Cell 128, 693–705 (2007). - PubMed
-
- Jenuwein T & Allis CD Translating the Histone Code. Science 293, 1074–1080 (2001). - PubMed
-
- Allis CD, Caparros ML, Jenuwein T & Reinberg D Epigenetics. (Cold Spring Harbor Laboratory Press, 2015).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

