Bacterial biofilms in infective endocarditis: an in vitro model to investigate emerging technologies of antimicrobial cardiovascular device coatings
- PMID: 32444905
- PMCID: PMC7907033
- DOI: 10.1007/s00392-020-01669-y
Bacterial biofilms in infective endocarditis: an in vitro model to investigate emerging technologies of antimicrobial cardiovascular device coatings
Abstract
Objective: In spite of the progress in antimicrobial and surgical therapy, infective endocarditis (IE) is still associated with a high morbidity and mortality. IE is characterized by bacterial biofilms of the endocardium, especially of the aortic and mitral valve leading to their destruction. About one quarter of patients with formal surgery indication cannot undergo surgery. This group of patients needs further options of therapy, but due to a lack of models for IE prospects of research are low. Therefore, the purpose of this project was to establish an in vitro model of infective endocarditis to allow growth of bacterial biofilms on porcine aortic valves, serving as baseline for further research.
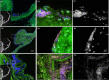
Methods and results: A pulsatile two-chamber circulation model was constructed that kept native porcine aortic valves under sterile, physiologic hemodynamic and temperature conditions. To create biofilms on porcine aortic valves the system was inoculated with Staphylococcus epidermidis PIA 8400. Aortic roots were incubated in the model for increasing periods of time (24 h and 40 h) and bacterial titration (1.5 × 104 CFU/mL and 1.5 × 105 CFU/mL) with 5 L cardiac output per minute. After incubation, tissue sections were analysed by fluorescence in situ hybridization (FISH) for direct visualization of the biofilms. Pilot tests for biofilm growth showed monospecies colonization consisting of cocci with time- and inocula-dependent increase after 24 h and 40 h (n = 4). In n = 3 experiments for 24 h, with the same inocula, FISH visualized biofilms with ribosome-containing, and thus metabolic active cocci, tissue infiltration and similar colonization pattern as observed by the FISH in human IE heart valves infected by S. epidermidis.
Conclusion: These results demonstrate the establishment of a novel in vitro model for bacterial biofilm growth on porcine aortic roots mimicking IE. The model will allow to identify predilection sites of valves for bacterial adhesion and biofilm growth and it may serve as baseline for further research on IE therapy and prevention, e.g. the development of antimicrobial transcatheter approaches to IE.
Keywords: Biofilm; Bioreactor; Fluorescence in situ hybridization; In vitro model; Infective endocarditis; Staphylococcus epidermidis.
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures



Similar articles
-
Associations between echocardiographic manifestations and bacterial species in patients with infective endocarditis: a cohort study.BMC Infect Dis. 2019 Dec 16;19(1):1052. doi: 10.1186/s12879-019-4682-z. BMC Infect Dis. 2019. PMID: 31842764 Free PMC article.
-
Rothia aeria and Rothia dentocariosa as biofilm builders in infective endocarditis.Int J Med Microbiol. 2021 Feb;311(2):151478. doi: 10.1016/j.ijmm.2021.151478. Epub 2021 Feb 6. Int J Med Microbiol. 2021. PMID: 33581548
-
Surgery for infective endocarditis: determinate factors in the outcome.J Cardiovasc Surg (Torino). 2008 Aug;49(4):545-8. J Cardiovasc Surg (Torino). 2008. PMID: 18665120
-
Infective endocarditis following transcatheter edge-to-edge mitral valve repair: A systematic review.Catheter Cardiovasc Interv. 2018 Sep 1;92(3):583-591. doi: 10.1002/ccd.27632. Epub 2018 May 10. Catheter Cardiovasc Interv. 2018. PMID: 29745455
-
Successful antimicrobial chemotherapy for nocardia asteroides prosthetic valve endocarditis.Am J Med. 2003 Sep;115(4):330-2. doi: 10.1016/s0002-9343(03)00350-4. Am J Med. 2003. PMID: 12967703 Review. No abstract available.
Cited by
-
Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models.iScience. 2021 Apr 17;24(5):102443. doi: 10.1016/j.isci.2021.102443. eCollection 2021 May 21. iScience. 2021. PMID: 34013169 Free PMC article. Review.
-
The Current Knowledge on the Pathogenesis of Tissue and Medical Device-Related Biofilm Infections.Microorganisms. 2022 Jun 21;10(7):1259. doi: 10.3390/microorganisms10071259. Microorganisms. 2022. PMID: 35888978 Free PMC article.
-
Anti-biofilm Approach in Infective Endocarditis Exposes New Treatment Strategies for Improved Outcome.Front Cell Dev Biol. 2021 Jun 18;9:643335. doi: 10.3389/fcell.2021.643335. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34222225 Free PMC article. Review.
-
Investigating the anti-streptococcal biofilm effect of ssDNA aptamer-silver nanoparticles complex on a titanium-based substrate.RSC Adv. 2022 Sep 15;12(38):24876-24886. doi: 10.1039/d2ra04112j. eCollection 2022 Aug 30. RSC Adv. 2022. PMID: 36276899 Free PMC article.
-
Strategies to Improve the Potency of Oxazolidinones towards Bacterial Biofilms.Chem Asian J. 2022 Jun 1;17(11):e202200201. doi: 10.1002/asia.202200201. Epub 2022 Apr 13. Chem Asian J. 2022. PMID: 35352479 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

