Phase-separated bacterial ribonucleoprotein bodies organize mRNA decay
- PMID: 32445438
- PMCID: PMC7554086
- DOI: 10.1002/wrna.1599
Phase-separated bacterial ribonucleoprotein bodies organize mRNA decay
Abstract
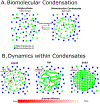
In bacteria, mRNA decay is controlled by megadalton scale macromolecular assemblies called, "RNA degradosomes," composed of nucleases and other RNA decay associated proteins. Recent advances in bacterial cell biology have shown that RNA degradosomes can assemble into phase-separated structures, termed bacterial ribonucleoprotein bodies (BR-bodies), with many analogous properties to eukaryotic processing bodies and stress granules. This review will highlight the functional role that BR-bodies play in the mRNA decay process through its organization into a membraneless organelle in the bacterial cytoplasm. This review will also highlight the phylogenetic distribution of BR-bodies across bacterial species, which suggests that these phase-separated structures are broadly distributed across bacteria, and in evolutionarily related mitochondria and chloroplasts. This article is categorized under: RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes RNA Export and Localization > RNA Localization RNA Turnover and Surveillance > Regulation of RNA Stability.
Keywords: RNA degradosome; bacterial ribonucleoprotein bodies; mRNA decay; phase separation.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no competing interests exist.
Figures


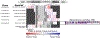


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

