Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets
- PMID: 32448084
- PMCID: PMC7448863
- DOI: 10.1080/22221751.2020.1772679
Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets
Abstract
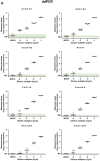
Different primers/probes sets have been developed all over the world for the nucleic acid detection of SARS-CoV-2 by quantitative real time polymerase chain reaction (qRT-PCR) as a standard method. In our recent study, we explored the feasibility of droplet digital PCR (ddPCR) for clinical SARS-CoV-2 nucleic acid detection compared with qRT-PCR using the same primer/probe sets issued by Chinese Center for Disease Control and Prevention (CDC) targeting viral ORF1ab or N gene, which showed that ddPCR could largely minimize the false negatives reports resulted by qRT-PCR [Suo T, Liu X, Feng J, et al. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv [Internet]. 2020;2020.02.29.20029439. Available from: https://medrxiv.org/content/early/2020/03/06/2020.02.29.20029439.abstract]. Here, we further stringently compared the performance of qRT-PCR and ddPCR for 8 primer/probe sets with the same clinical samples and conditions. Results showed that none of 8 primer/probe sets used in qRT-PCR could significantly distinguish true negatives and positives with low viral load (10-4 dilution). Moreover, false positive reports of qRT-PCR with UCDC-N1, N2 and CCDC-N primers/probes sets were observed. In contrast, ddPCR showed significantly better performance in general for low viral load samples compared to qRT-PCR. Remarkably, the background readouts of ddPCR are relatively lower, which could efficiently reduce the production of false positive reports.
Keywords: SARS-CoV-2; diagnosis; digital PCR; false negative; false positive; real time PCR.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Suo T, Liu X, Feng J, et al. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv [Internet]. 2020;2020.02.29.20029439. Available from: https://medrxiv.org/content/early/2020/03/06/2020.02.29.20029439.abstract. - PMC - PubMed
-
- Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv [Internet]. 2020;2020.03.30.20048108. Available from: https://medrxiv.org/content/early/2020/04/01/2020.03.30.20048108.abstract.
-
- National Institute for Viral Disease Control and Prevention of PRC. Specific primers and probes for detection 2019 novel coronavirus [Internet]. 2020 [cited 2020 Apr 10]. Available from: https://www.chinaivdc.cn/kyjz/202001/t20200121_211337.html.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
