Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor
- PMID: 32448098
- PMCID: PMC7284149
- DOI: 10.1080/07391102.2020.1773318
Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor
Abstract
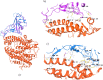
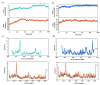
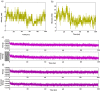
The novel corona virus disease 2019 (SARS-CoV 2) pandemic outbreak was alarming. The binding of SARS-CoV (CoV) spike protein (S-Protein) Receptor Binding Domain (RBD) to Angiotensin converting enzyme 2 (ACE2) receptor initiates the entry of corona virus into the host cells leading to the infection. However, considering the mutations reported in the SARS-CoV 2 (nCoV), the structural changes and the binding interactions of the S-protein RBD of nCoV were not clear. The present study was designed to elucidate the structural changes, hot spot binding residues and their interactions between the nCoV S-protein RBD and ACE2 receptor through computational approaches. Based on the sequence alignment, a total of 58 residues were found mutated in nCoV S-protein RBD. These mutations led to the structural changes in the nCoV S-protein RBD 3d structure with 4 helices, 10 sheets and intermittent loops. The nCoV RBD was found binding to ACE2 receptor with 11 hydrogen bonds and 1 salt bridge. The major hot spot amino acids involved in the binding identified by interaction analysis after simulations includes Glu 35, Tyr 83, Asp 38, Lys 31, Glu 37, His 34 amino acid residues of ACE2 receptor and Gln 493, Gln 498, Asn 487, Tyr 505 and Lys 417 residues in nCoV S-protein RBD. Based on the hydrogen bonding, RMSD and RMSF, total and potential energies, the nCoV was found binding to ACE2 receptor with higher stability and rigidity. Concluding, the hotspots information will be useful in designing blockers for the nCoV spike protein RBD. [Formula: see text]Communicated by Ramaswamy H. Sarma.
Keywords: ACE2 receptor; Novel Coronavirus; hotspots; molecular dynamic simulations; protein interactions; receptor binding domain; spike protein.
Figures

Similar articles
-
Effect of mutation on structure, function and dynamics of receptor binding domain of human SARS-CoV-2 with host cell receptor ACE2: a molecular dynamics simulations study.J Biomol Struct Dyn. 2021 Nov;39(18):7231-7245. doi: 10.1080/07391102.2020.1802348. Epub 2020 Aug 7. J Biomol Struct Dyn. 2021. PMID: 32762417 Free PMC article.
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Probing structural basis for enhanced binding of SARS-CoV-2 P.1 variant spike protein with the human ACE2 receptor.J Cell Biochem. 2022 Jul;123(7):1207-1221. doi: 10.1002/jcb.30276. Epub 2022 May 27. J Cell Biochem. 2022. PMID: 35620980 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
Cited by
-
Biomolecular interactions with nanoparticles: applications for coronavirus disease 2019.Curr Opin Colloid Interface Sci. 2021 Aug;54:101461. doi: 10.1016/j.cocis.2021.101461. Epub 2021 Apr 23. Curr Opin Colloid Interface Sci. 2021. PMID: 33907504 Free PMC article. Review.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants.Cell Mol Life Sci. 2021 Dec;78(24):7967-7989. doi: 10.1007/s00018-021-04008-0. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34731254 Free PMC article. Review.
-
Computational Modeling of T Cell Hypersensitivity during Coronavirus Infections Leading to Autoimmunity and Lethality.Comput Math Methods Med. 2022 Mar 22;2022:9444502. doi: 10.1155/2022/9444502. eCollection 2022. Comput Math Methods Med. 2022. PMID: 35341005 Free PMC article.
-
Small molecule therapeutics to destabilize the ACE2-RBD complex: A molecular dynamics study.Biophys J. 2021 Jul 20;120(14):2793-2804. doi: 10.1016/j.bpj.2021.06.016. Epub 2021 Jun 30. Biophys J. 2021. PMID: 34214539 Free PMC article.
References
-
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics (Just-Accepted), 1–17. 10.1080/07391102.2020.1763199 - DOI - PMC - PubMed
-
- Adeoye A. O., Oso B. J., Olaoye I. F., Tijjani H., & Adebayo A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics (Just-Accepted), 1–14. 10.1080/07391102.2020.1765876 - DOI - PMC - PubMed
-
- Aydin H., Al‐Khooly D., & Lee J. E. (2014). Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: insights into mechanisms of general viral fusion and inhibitor design. Protein Science: A Publication of the Protein Society, 23(5), 603–617. 10.1002/pro.2442 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
