Bio-semantic relation extraction with attention-based external knowledge reinforcement
- PMID: 32448122
- PMCID: PMC7245897
- DOI: 10.1186/s12859-020-3540-8
Bio-semantic relation extraction with attention-based external knowledge reinforcement
Abstract
Background: Semantic resources such as knowledge bases contains high-quality-structured knowledge and therefore require significant effort from domain experts. Using the resources to reinforce the information retrieval from the unstructured text may further exploit the potentials of such unstructured text resources and their curated knowledge.
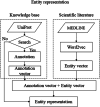
Results: The paper proposes a novel method that uses a deep neural network model adopting the prior knowledge to improve performance in the automated extraction of biological semantic relations from the scientific literature. The model is based on a recurrent neural network combining the attention mechanism with the semantic resources, i.e., UniProt and BioModels. Our method is evaluated on the BioNLP and BioCreative corpus, a set of manually annotated biological text. The experiments demonstrate that the method outperforms the current state-of-the-art models, and the structured semantic information could improve the result of bio-text-mining.
Conclusion: The experiment results show that our approach can effectively make use of the external prior knowledge information and improve the performance in the protein-protein interaction extraction task. The method should be able to be generalized for other types of data, although it is validated on biomedical texts.
Keywords: Attention mechanism; Bio-text-mining; Biological semantic relation; Knowledge base.
Conflict of interest statement
The authors have no conflict of interest related to the research presented herein.
Figures




Similar articles
-
Chemical-induced disease relation extraction with dependency information and prior knowledge.J Biomed Inform. 2018 Aug;84:171-178. doi: 10.1016/j.jbi.2018.07.007. Epub 2018 Jul 11. J Biomed Inform. 2018. PMID: 30017973
-
Biomedical event causal relation extraction with deep knowledge fusion and Roberta-based data augmentation.Methods. 2024 Nov;231:8-14. doi: 10.1016/j.ymeth.2024.08.007. Epub 2024 Sep 4. Methods. 2024. PMID: 39241919
-
Neural sentence embedding models for semantic similarity estimation in the biomedical domain.BMC Bioinformatics. 2019 Apr 11;20(1):178. doi: 10.1186/s12859-019-2789-2. BMC Bioinformatics. 2019. PMID: 30975071 Free PMC article.
-
Path-based knowledge reasoning with textual semantic information for medical knowledge graph completion.BMC Med Inform Decis Mak. 2021 Nov 29;21(Suppl 9):335. doi: 10.1186/s12911-021-01622-7. BMC Med Inform Decis Mak. 2021. PMID: 34844576 Free PMC article. Review.
-
Enriching contextualized language model from knowledge graph for biomedical information extraction.Brief Bioinform. 2021 May 20;22(3):bbaa110. doi: 10.1093/bib/bbaa110. Brief Bioinform. 2021. PMID: 32591802 Review.
Cited by
-
Lisen&Curate: A platform to facilitate gathering textual evidence for curation of regulation of transcription initiation in bacteria.Biochim Biophys Acta Gene Regul Mech. 2021 Nov-Dec;1864(11-12):194753. doi: 10.1016/j.bbagrm.2021.194753. Epub 2021 Aug 28. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 34461312 Free PMC article.
References
-
- Chelliah V, Laibe C, Novère NL. Encyclopedia of Systems Biology. New York, NY: Springer; 2013. BioModels database: a repository of mathematical models of biological processes. - PubMed
-
- Krallinger M, Vazquez M, Leitner F, Salgado D, Chatr-Aryamontri A, Winter A, Perfetto L, Briganti L, Licata L, Iannuccelli M, Castagnoli L, Cesareni G, Tyers M, Schneider G, Rinaldi F, Leaman R, Gonzalez G, Matos S, Kim S, Wilbur WJ, Rocha L, Shatkay H, Tendulkar AV, Agarwal S, Liu F, Wang X, Rak R, Noto K, Elkan C, Lu Z, Dogan RI, Fontaine JF, Andrade-Navarro MA, Valencia A. The protein-protein interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinformatics. 2011;12(8):S3. doi: 10.1186/1471-2105-12-S8-S3. - DOI - PMC - PubMed
-
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2015;122:957–968. doi: 10.1016/j.cell.2005.08.029. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

