LRIG1 gene copy number analysis by ddPCR and correlations to clinical factors in breast cancer
- PMID: 32448168
- PMCID: PMC7245921
- DOI: 10.1186/s12885-020-06919-w
LRIG1 gene copy number analysis by ddPCR and correlations to clinical factors in breast cancer
Abstract
Background: Leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) copy number alterations and unbalanced gene recombination events have been reported to occur in breast cancer. Importantly, LRIG1 loss was recently shown to predict early and late relapse in stage I-II breast cancer.
Methods: We developed droplet digital PCR (ddPCR) assays for the determination of relative LRIG1 copy numbers and used these assays to analyze LRIG1 in twelve healthy individuals, 34 breast tumor samples previously analyzed by fluorescence in situ hybridization (FISH), and 423 breast tumor cytosols.
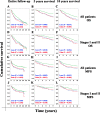
Results: Four of the LRIG1/reference gene assays were found to be precise and robust, showing copy number ratios close to 1 (mean, 0.984; standard deviation, +/- 0.031) among the healthy control population. The correlation between the ddPCR assays and previous FISH results was low, possibly because of the different normalization strategies used. One in 34 breast tumors (2.9%) showed an unbalanced LRIG1 recombination event. LRIG1 copy number ratios were associated with the breast cancer subtype, steroid receptor status, ERBB2 status, tumor grade, and nodal status. Both LRIG1 loss and gain were associated with unfavorable metastasis-free survival; however, they did not remain significant prognostic factors after adjustment for common risk factors in the Cox regression analysis. Furthermore, LRIG1 loss was not significantly associated with survival in stage I and II cases.
Conclusions: Although LRIG1 gene aberrations may be important determinants of breast cancer biology, and prognostic markers, the results of this study do not verify an important role for LRIG1 copy number analyses in predicting the risk of relapse in early-stage breast cancer.
Keywords: Breast cancer; Gene copy number; LRIG1; Prognosis; ddPCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

