Structural and functional similarities and differences in nucleolar Pumilio RNA-binding proteins between Arabidopsis and the charophyte Chara corallina
- PMID: 32448230
- PMCID: PMC7247198
- DOI: 10.1186/s12870-020-02444-x
Structural and functional similarities and differences in nucleolar Pumilio RNA-binding proteins between Arabidopsis and the charophyte Chara corallina
Abstract
Background: Pumilio RNA-binding proteins are evolutionarily conserved throughout eukaryotes and are involved in RNA decay, transport, and translation repression in the cytoplasm. Although a majority of Pumilio proteins function in the cytoplasm, two nucleolar forms have been reported to have a function in rRNA processing in Arabidopsis. The species of the genus Chara have been known to be most closely related to land plants, as they share several characteristics with modern Embryophyta.
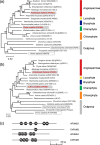
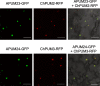
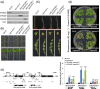
Results: In this study, we identified two putative nucleolar Pumilio protein genes, namely, ChPUM2 and ChPUM3, from the transcriptome of Chara corallina. Of the two ChPUM proteins, ChPUM2 was most similar in amino acid sequence (27% identity and 45% homology) and predicted protein structure to Arabidopsis APUM23, while ChPUM3 was similar to APUM24 (35% identity and 54% homology). The transient expression of 35S:ChPUM2-RFP and 35S:ChPUM3-RFP showed nucleolar localization of fusion proteins in tobacco leaf cells, similar to the expression of 35S:APUM23-GFP and 35S:APUM24-GFP. Moreover, 35S:ChPUM2 complemented the morphological defects of the apum23 phenotypes but not those of apum24, while 35S:ChPUM3 could not complement the apum23 and apum24 mutants. Similarly, the 35S:ChPUM2/apum23 plants rescued the pre-rRNA processing defect of apum23, but 35S:ChPUM3/apum24+/- plants did not rescue that of apum24. Consistent with these complementation results, a known target RNA-binding sequence at the end of the 18S rRNA (5'-GGAAUUGACGG) for APUM23 was conserved in Arabidopsis and C. corallina, whereas a target region of ITS2 pre-rRNA for APUM24 was 156 nt longer in C. corallina than in A. thaliana. Moreover, ChPUM2 and APUM23 were predicted to have nearly identical structures, but ChPUM3 and APUM24 have different structures in the 5th C-terminal Puf RNA-binding domain, which had a longer random coil in ChPUM3 than in APUM24.
Conclusions: ChPUM2 of C. corallina was functional in Arabidopsis, similar to APUM23, but ChPUM3 did not substitute for APUM24 in Arabidopsis. Protein homology modeling showed high coverage between APUM23 and ChPUM2, but displayed structural differences between APUM24 and ChPUM3. Together with the protein structure of ChPUM3 itself, a short ITS2 of Arabidopsis pre-rRNA may interrupt the binding of ChPUM3 to 3'-extended 5.8S pre-rRNA.
Keywords: Arabidopsis thaliana; Chara corallina; Charophyta; ITS2; Puf; RNA-binding proteins; rRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis.Plant J. 2010 Dec;64(6):960-76. doi: 10.1111/j.1365-313X.2010.04393.x. Epub 2010 Nov 17. Plant J. 2010. PMID: 21143677
-
An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing.Plant J. 2017 Dec;92(6):1092-1105. doi: 10.1111/tpj.13745. Epub 2017 Nov 20. Plant J. 2017. PMID: 29031033
-
Reduced Expression of APUM24, Encoding a Novel rRNA Processing Factor, Induces Sugar-Dependent Nucleolar Stress and Altered Sugar Responses in Arabidopsis thaliana.Plant Cell. 2018 Jan;30(1):209-227. doi: 10.1105/tpc.17.00778. Epub 2017 Dec 14. Plant Cell. 2018. PMID: 29242314 Free PMC article.
-
Pumilio Puf domain RNA-binding proteins in Arabidopsis.Plant Signal Behav. 2011 Mar;6(3):364-8. doi: 10.4161/psb.6.3.14380. Epub 2011 Mar 1. Plant Signal Behav. 2011. PMID: 21350339 Free PMC article. Review.
-
Cell and molecular biology of the fastest myosins.Int Rev Cell Mol Biol. 2009;276:301-47. doi: 10.1016/S1937-6448(09)76007-1. Int Rev Cell Mol Biol. 2009. PMID: 19584016 Review.
Cited by
-
Rediscovering Chara as a model organism for molecular and evo-devo studies.Protoplasma. 2024 Mar;261(2):183-196. doi: 10.1007/s00709-023-01900-3. Epub 2023 Oct 25. Protoplasma. 2024. PMID: 37880545 Review.
-
Comprehensive Identification of the Pum Gene Family and Its Involvement in Kernel Development in Maize.Int J Mol Sci. 2023 Sep 13;24(18):14036. doi: 10.3390/ijms241814036. Int J Mol Sci. 2023. PMID: 37762337 Free PMC article.
-
The Role of Pumilio RNA Binding Protein in Plants.Biomolecules. 2021 Dec 9;11(12):1851. doi: 10.3390/biom11121851. Biomolecules. 2021. PMID: 34944494 Free PMC article. Review.
-
Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview.Int J Mol Sci. 2021 Jun 23;22(13):6731. doi: 10.3390/ijms22136731. Int J Mol Sci. 2021. PMID: 34201749 Free PMC article. Review.
References
-
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. - PubMed
-
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. - PubMed
-
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. - PubMed
-
- Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

