Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions
- PMID: 32448240
- PMCID: PMC7245778
- DOI: 10.1186/s12915-020-00782-8
Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions
Abstract
Background: Dinoflagellates are taxonomically diverse and ecologically important phytoplankton that are ubiquitously present in marine and freshwater environments. Mostly photosynthetic, dinoflagellates provide the basis of aquatic primary production; most taxa are free-living, while some can form symbiotic and parasitic associations with other organisms. However, knowledge of the molecular mechanisms that underpin the adaptation of these organisms to diverse ecological niches is limited by the scarce availability of genomic data, partly due to their large genome sizes estimated up to 250 Gbp. Currently available dinoflagellate genome data are restricted to Symbiodiniaceae (particularly symbionts of reef-building corals) and parasitic lineages, from taxa that have smaller genome size ranges, while genomic information from more diverse free-living species is still lacking.
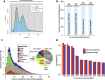
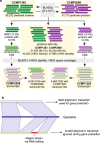
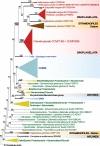
Results: Here, we present two draft diploid genome assemblies of the free-living dinoflagellate Polarella glacialis, isolated from the Arctic and Antarctica. We found that about 68% of the genomes are composed of repetitive sequence, with long terminal repeats likely contributing to intra-species structural divergence and distinct genome sizes (3.0 and 2.7 Gbp). For each genome, guided using full-length transcriptome data, we predicted > 50,000 high-quality protein-coding genes, of which ~40% are in unidirectional gene clusters and ~25% comprise single exons. Multi-genome comparison unveiled genes specific to P. glacialis and a common, putatively bacterial origin of ice-binding domains in cold-adapted dinoflagellates.
Conclusions: Our results elucidate how selection acts within the context of a complex genome structure to facilitate local adaptation. Because most dinoflagellate genes are constitutively expressed, Polarella glacialis has enhanced transcriptional responses via unidirectional, tandem duplication of single-exon genes that encode functions critical to survival in cold, low-light polar environments. These genomes provide a foundational reference for future research on dinoflagellate evolution.
Keywords: Cold adaptation; Dinoflagellates; Genome evolution; Genomics; Polarella glacialis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium.BMC Biol. 2021 Apr 13;19(1):73. doi: 10.1186/s12915-021-00994-6. BMC Biol. 2021. PMID: 33849527 Free PMC article.
-
Gene duplication is the primary driver of intraspecific genomic divergence in coral algal symbionts.Open Biol. 2023 Sep;13(9):230182. doi: 10.1098/rsob.230182. Epub 2023 Sep 27. Open Biol. 2023. PMID: 37751888 Free PMC article.
-
Massive genome reduction predates the divergence of Symbiodiniaceae dinoflagellates.ISME J. 2024 Jan 8;18(1):wrae059. doi: 10.1093/ismejo/wrae059. ISME J. 2024. PMID: 38655774 Free PMC article.
-
Genome Evolution of Coral Reef Symbionts as Intracellular Residents.Trends Ecol Evol. 2019 Sep;34(9):799-806. doi: 10.1016/j.tree.2019.04.010. Epub 2019 May 10. Trends Ecol Evol. 2019. PMID: 31084944 Review.
-
Sex in Symbiodiniaceae dinoflagellates: genomic evidence for independent loss of the canonical synaptonemal complex.Sci Rep. 2020 Jun 17;10(1):9792. doi: 10.1038/s41598-020-66429-4. Sci Rep. 2020. PMID: 32555361 Free PMC article. Review.
Cited by
-
Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium.BMC Biol. 2021 Apr 13;19(1):73. doi: 10.1186/s12915-021-00994-6. BMC Biol. 2021. PMID: 33849527 Free PMC article.
-
Genome-wide identification, comparative analysis and functional roles in flavonoid biosynthesis of cytochrome P450 superfamily in pear (Pyrus spp.).BMC Genom Data. 2023 Oct 3;24(1):58. doi: 10.1186/s12863-023-01159-w. BMC Genom Data. 2023. PMID: 37789271 Free PMC article.
-
Evidence That Inconsistent Gene Prediction Can Mislead Analysis of Dinoflagellate Genomes.J Phycol. 2020 Feb;56(1):6-10. doi: 10.1111/jpy.12947. J Phycol. 2020. PMID: 31713873 Free PMC article.
-
Gene clusters for biosynthesis of mycosporine-like amino acids in dinoflagellate nuclear genomes: Possible recent horizontal gene transfer between species of Symbiodiniaceae (Dinophyceae).J Phycol. 2022 Feb;58(1):1-11. doi: 10.1111/jpy.13219. Epub 2021 Nov 26. J Phycol. 2022. PMID: 34699617 Free PMC article. Review.
-
Extreme environments offer an unprecedented opportunity to understand microbial eukaryotic ecology, evolution, and genome biology.Nat Commun. 2023 Aug 16;14(1):4959. doi: 10.1038/s41467-023-40657-4. Nat Commun. 2023. PMID: 37587119 Free PMC article. Review.
References
-
- Guiry MD. How many species of algae are there? J Phycol. 2012;48(5):1057–1063. - PubMed
-
- Fensome RA, MacRae RA, Moldowan JM, Taylor FJR, Williams GL. The early Mesozoic radiation of dinoflagellates. Paleobiology. 2016;22(3):329–338.
-
- Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst. 2003;34(1):661–689.
-
- LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 2018;28(16):2570–2580. - PubMed
-
- Stentiford GD, Shields JD. A review of the parasitic dinoflagellates Hematodinium species and Hematodinium-like infections in marine crustaceans. Dis Aquat Org. 2005;66(1):47–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

