Fifteen new nucleotide substitutions in variants of human papillomavirus 18 in Korea : Korean HPV18 variants and clinical manifestation
- PMID: 32448303
- PMCID: PMC7245819
- DOI: 10.1186/s12985-020-01337-7
Fifteen new nucleotide substitutions in variants of human papillomavirus 18 in Korea : Korean HPV18 variants and clinical manifestation
Abstract
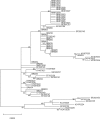
High-risk human papillomavirus (HPV) infection is an essential factor for the development of cervical cancer. HPV18 is the second most common carcinogenic HPV type following HPV16, but the lineages of HPV18 have been less well studied than those of HPV 16. The purpose of this study was to analyze the nucleotide variants in the E6, E7, and L1 genes of HPV18, to assess the prevalence of HPV18 variants in Korea and to explore the relationship between HPV18 genetic variants and the risk for cervical cancer.A total of 170 DNA samples from HPV18-positive cervical specimens were collected from women admitted to a secondary referral hospital located in Seoul. Among them, the lineages of the 97 samples could be successfully determined by historical nomenclature.All the studied HPV 18 variants were lineage A. Sublineages A1 and A4 comprised 91.7% (89/97) and 1.0% (1/97), respectively. Sublineages other than A1 or A4 comprised 7.2% (7/97). We identified 15 new nucleotide substitutions among 44 nucleotide substitutions: C158T, T317G, T443G, A560G, A5467G, A5560C, A5678C, A6155G, G6462A, T6650G, G6701A, T6809C, A6823G, T6941C and T6953C. Among them, 6 substitutions at positions 317, 443, 5467, 5560, 6462, and 6823 resulted in amino acid changes (E6: F71L and N113K; L1: H13R, H44P, A345T, and N465S, respectively). The pathologic results were classified as normal in 25.8% (25/97) of the women, atypical squamous cells of undermined significance (ASCUS) in 7.2% (7/97), cervical intraepithelial neoplasia (CIN) 1 in 36.1% (35/97), CIN2/3 in 19.6% (18/97), and carcinoma in 12.4% (12/97). There was no significant association between the HPV18 sublineages and the severity of pathologic lesion or the disease progression.This study is the first to analyze the distribution of HPV18 variants in Korean and to associate the results with pathologic findings. Although the HPV18 variants had no significant effect on the degree and progression of the disease, the newly discovered nonsynonymous mutation in L1 might serve as a database to determine vaccine efficacy in Korean women.
Keywords: Cervical cancer; E6, E7 and L1 genes; Human papillomavirus (HPV) 18; Lineage; Variants.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Genetic variability of human papillomavirus type 18 based on E6, E7 and L1 genes in central China.Virol J. 2024 Jul 5;21(1):152. doi: 10.1186/s12985-024-02424-9. Virol J. 2024. PMID: 38970084 Free PMC article.
-
Human papillomavirus (HPV) 18 genetic variants and cervical cancer risk in Taizhou area, China.Gene. 2018 Mar 20;647:192-197. doi: 10.1016/j.gene.2018.01.037. Epub 2018 Jan 10. Gene. 2018. PMID: 29331480
-
Phylogeny and Classification of Human Papillomavirus (HPV)16 and HPV18 Variants Based on E6 and L1 genes in Tunisian Women with Cervical Lesions.Asian Pac J Cancer Prev. 2018 Dec 25;19(12):3361-3366. doi: 10.31557/APJCP.2018.19.12.3361. Asian Pac J Cancer Prev. 2018. PMID: 30583341 Free PMC article.
-
Association of human papillomavirus type 16 and its genetic variants with cervical lesion in Korea.APMIS. 2016 Nov;124(11):950-957. doi: 10.1111/apm.12592. Epub 2016 Aug 21. APMIS. 2016. PMID: 27546189
-
Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study.Lancet Oncol. 2011 Sep;12(9):880-90. doi: 10.1016/S1470-2045(11)70188-7. Epub 2011 Aug 22. Lancet Oncol. 2011. PMID: 21865084 Clinical Trial.
Cited by
-
Type distribution of human papillomaviruses in ThinPrep cytology samples and HPV16/18 E6 gene variations in FFPE cervical cancer specimens in Fars province, Iran.Cancer Cell Int. 2023 Aug 11;23(1):166. doi: 10.1186/s12935-023-03011-8. Cancer Cell Int. 2023. PMID: 37568237 Free PMC article.
-
Determination of Human Papillomavirus Type 18 Lineage of E6: A Population Study from Iran.Biomed Res Int. 2022 Mar 18;2022:2839708. doi: 10.1155/2022/2839708. eCollection 2022. Biomed Res Int. 2022. PMID: 35342765 Free PMC article.
-
Distribution of human papillomavirus genotypes in suspected women cytological specimens from Tehran, Iran.Iran J Microbiol. 2022 Feb;14(1):112-118. doi: 10.18502/ijm.v14i1.8812. Iran J Microbiol. 2022. PMID: 35664716 Free PMC article.
-
Genetic variability of human papillomavirus type 18 based on E6, E7 and L1 genes in central China.Virol J. 2024 Jul 5;21(1):152. doi: 10.1186/s12985-024-02424-9. Virol J. 2024. PMID: 38970084 Free PMC article.
References
-
- WHO Global Cancer Observatory [Available from: https://gco.iarc.fr/].
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

