Identification of a new effector-immunity pair of Aeromonas hydrophila type VI secretion system
- PMID: 32448355
- PMCID: PMC7245790
- DOI: 10.1186/s13567-020-00794-w
Identification of a new effector-immunity pair of Aeromonas hydrophila type VI secretion system
Abstract
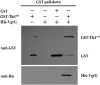
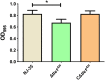
The type VI secretion system (T6SS) is a multiprotein weapon that kills eukaryotic predators or prokaryotic competitors by delivering toxic effectors. Despite the importance of T6SS in bacterial environmental adaptation, it is still challenging to systematically identify T6SS effectors because of their high diversity and lack of conserved domains. In this report, we discovered a putative effector gene, U876-17730, in the whole genome of Aeromonas hydrophila NJ-35 based on the reported conservative domain DUF4123 (domain of unknown function), with two cognate immunity proteins encoded downstream. Phylogenetic tree analysis of amino acids indicates that AH17730 belongs to the Tle1 (type VI lipase effector) family, and therefore was named Tle1AH. The deletion of tle1AH resulted in significantly decreased biofilm formation, antibacterial competition ability and virulence in zebrafish (Danio rerio) when compared to the wild-type strain. Only when the two immunity proteins coexist can bacteria protect themselves from the toxicity of Tle1AH. Further study shows that Tle1AH is a kind of phospholipase that possesses a conserved lipase motif, Gly-X-Ser-X-Gly (X is for any amino acid). Tle1AH is secreted by T6SS, and this secretion requires its interaction with an associated VgrG (valine-glycine repeat protein G). In conclusion, we identified a T6SS effector-immunity pair and verified its function, which lays the foundation for future research on the role of T6SS in the pathogenic mechanism of A. hydrophila.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis.Microbiology (Reading). 2013 Jun;159(Pt 6):1120-1135. doi: 10.1099/mic.0.063495-0. Epub 2013 Mar 21. Microbiology (Reading). 2013. PMID: 23519162 Free PMC article.
-
Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila.Microb Pathog. 2008 Apr;44(4):344-61. doi: 10.1016/j.micpath.2007.10.005. Epub 2007 Oct 24. Microb Pathog. 2008. PMID: 18037263 Free PMC article.
-
Identification of divergent type VI secretion effectors using a conserved chaperone domain.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):9106-11. doi: 10.1073/pnas.1505317112. Epub 2015 Jul 6. Proc Natl Acad Sci U S A. 2015. PMID: 26150500 Free PMC article.
-
The Type VI secretion system: a versatile bacterial weapon.Microbiology (Reading). 2019 May;165(5):503-515. doi: 10.1099/mic.0.000789. Epub 2019 Mar 20. Microbiology (Reading). 2019. PMID: 30893029 Review.
-
Type VI Secretion Effectors: Methodologies and Biology.Front Cell Infect Microbiol. 2017 Jun 15;7:254. doi: 10.3389/fcimb.2017.00254. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28664151 Free PMC article. Review.
Cited by
-
A novel chaperone-effector-immunity system identified in uropathogenic Escherichia coli UMN026.PeerJ. 2024 May 20;12:e17336. doi: 10.7717/peerj.17336. eCollection 2024. PeerJ. 2024. PMID: 38784397 Free PMC article.
-
From insect endosymbiont to phloem colonizer: comparative genomics unveils the lifestyle transition of phytopathogenic Arsenophonus strains.mSystems. 2025 May 20;10(5):e0149624. doi: 10.1128/msystems.01496-24. Epub 2025 Apr 9. mSystems. 2025. PMID: 40202301 Free PMC article.
-
Role of nasal microbiota in regulating host anti-influenza immunity in dogs.Microbiome. 2025 Jan 27;13(1):27. doi: 10.1186/s40168-025-02031-y. Microbiome. 2025. PMID: 39871363 Free PMC article.
-
Fur functions as an activator of T6SS-mediated bacterial dominance and virulence in Aeromonas hydrophila.Front Microbiol. 2023 Feb 8;13:1099611. doi: 10.3389/fmicb.2022.1099611. eCollection 2022. Front Microbiol. 2023. PMID: 36845974 Free PMC article.
-
A Global Survey of Hypervirulent Aeromonas hydrophila (vAh) Identified vAh Strains in the Lower Mekong River Basin and Diverse Opportunistic Pathogens from Farmed Fish and Other Environmental Sources.Microbiol Spectr. 2023 Feb 23;11(2):e0370522. doi: 10.1128/spectrum.03705-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36815836 Free PMC article.
References
-
- Pemberton JM, Kidd SP, Schmidt R. Secreted enzymes of Aeromonas. FEMS Microbiol Lett. 1997;152:1–10. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

