Marked Up-Regulation of ACE2 in Hearts of Patients With Obstructive Hypertrophic Cardiomyopathy: Implications for SARS-CoV-2-Mediated COVID-19
- PMID: 32448590
- PMCID: PMC7186205
- DOI: 10.1016/j.mayocp.2020.04.028
Marked Up-Regulation of ACE2 in Hearts of Patients With Obstructive Hypertrophic Cardiomyopathy: Implications for SARS-CoV-2-Mediated COVID-19
Abstract
Objective: To explore the transcriptomic differences between patients with hypertrophic cardiomyopathy (HCM) and controls.
Patients and methods: RNA was extracted from cardiac tissue flash frozen at therapeutic surgical septal myectomy for 106 patients with HCM and 39 healthy donor hearts. Expression profiling of 37,846 genes was performed using the Illumina Human HT-12v3 Expression BeadChip. All patients with HCM were genotyped for pathogenic variants causing HCM. Technical validation was performed using quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. This study was started on January 1, 1999, and final analysis was completed on April 20, 2020.
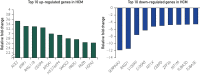
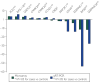
Results: Overall, 22% of the transcriptome (8443 of 37,846 genes) was expressed differentially between HCM and control tissues. Analysis by genotype revealed that gene expression changes were similar among genotypic subgroups of HCM, with only 4% (1502 of 37,846) to 6% (2336 of 37,846) of the transcriptome exhibiting differential expression between genotypic subgroups. The qRT-PCR confirmed differential expression in 92% (11 of 12 genes) of tested transcripts. Notably, in the context of coronavirus disease 2019 (COVID-19), the transcript for angiotensin I converting enzyme 2 (ACE2), a negative regulator of the angiotensin system, was the single most up-regulated gene in HCM (fold-change, 3.53; q-value =1.30×10-23), which was confirmed by qRT-PCR in triplicate (fold change, 3.78; P=5.22×10-4), and Western blot confirmed greater than 5-fold overexpression of ACE2 protein (fold change, 5.34; P=1.66×10-6).
Conclusion: More than 20% of the transcriptome is expressed differentially between HCM and control tissues. Importantly, ACE2 was the most up-regulated gene in HCM, indicating perhaps the heart's compensatory effort to mount an antihypertrophic, antifibrotic response. However, given that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) uses ACE2 for viral entry, this 5-fold increase in ACE2 protein may confer increased risk for COVID-19 manifestations and outcomes in patients with increased ACE2 transcript expression and protein levels in the heart.
Copyright © 2020 Mayo Foundation for Medical Education and Research. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Maron B.J., Gardin J.M., Flack J.M., Gidding S.S., Kurosaki T.T., Bild D.E. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. - PubMed
-
- Maron B.J., Roberts W.C., McAllister H.A., Rosing D.R., Epstein S.E. Sudden death in young athletes. Circulation. 1980;62(2):218–229. - PubMed
-
- Poetter K., Jiang H., Hassanzadeh S. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13(1):63–69. - PubMed
-
- Morner S., Richard P., Kazzam E. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. J Mol Cell Cardiol. 2003;35(7):841–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

