Evolution of the Insecticide Target Rdl in African Anopheles Is Driven by Interspecific and Interkaryotypic Introgression
- PMID: 32449755
- PMCID: PMC7530614
- DOI: 10.1093/molbev/msaa128
Evolution of the Insecticide Target Rdl in African Anopheles Is Driven by Interspecific and Interkaryotypic Introgression
Abstract
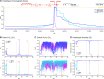
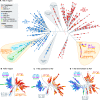
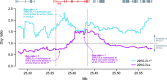
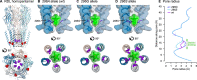
The evolution of insecticide resistance mechanisms in natural populations of Anopheles malaria vectors is a major public health concern across Africa. Using genome sequence data, we study the evolution of resistance mutations in the resistance to dieldrin locus (Rdl), a GABA receptor targeted by several insecticides, but most notably by the long-discontinued cyclodiene, dieldrin. The two Rdl resistance mutations (296G and 296S) spread across West and Central African Anopheles via two independent hard selective sweeps that included likely compensatory nearby mutations, and were followed by a rare combination of introgression across species (from A. gambiae and A. arabiensis to A. coluzzii) and across nonconcordant karyotypes of the 2La chromosomal inversion. Rdl resistance evolved in the 1950s as the first known adaptation to a large-scale insecticide-based intervention, but the evolutionary lessons from this system highlight contemporary and future dangers for management strategies designed to combat development of resistance in malaria vectors.
Keywords: insect vectors; insecticide resistance; population genomics.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





References
-
- Anopheles gambiae 1000 Genomes Consortium. 2017. Ag1000G phase 2 AR1 data release. MalariaGEN [Internet]. Available from: https://www.malariagen.net/data/ag1000g-phase-2-ar1. Accessed June 6, 2020.
-
- Andolfatto P, Depaulis F, Navarro A.. 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 77(1):1–8. - PubMed
-
- Asahi M, Kobayashi M, Matsui H, Nakahira K.. 2015. Differential mechanisms of action of the novel γ-aminobutyric acid receptor antagonist ectoparasiticides fluralaner (A1443) and fipronil. Pest Manag Sci. 71(1):91–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

