LipF increases rifampicin and streptomycin sensitivity in a Mycobacterium tuberculosis surrogate
- PMID: 32450809
- PMCID: PMC7249682
- DOI: 10.1186/s12866-020-01802-x
LipF increases rifampicin and streptomycin sensitivity in a Mycobacterium tuberculosis surrogate
Abstract
Background: Mortality due to tuberculosis (TB) has increased due to the development of drug resistance, the mechanisms of which have not been fully elucidated. Our research group identified a low expression of lipF gene in Mycobacterium tuberculosis clinical isolates with drug resistance. The aim of this work was to evaluate the effect of lipase F (LipF) expression on mycobacterial drug resistance.
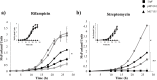
Results: The effects of expressing lipF from Mycobacterium tuberculosis in Mycobacterium smegmatis on resistance to antituberculosis drugs were determined with resazurin microtiter assay plate and growth kinetics. Functionality of ectopic LipF was confirmed. LipF expression reduced the rifampicin (RIF) and streptomycin (STR) minimum inhibitory concentration (MIC) from 3.12 μg/mL to 1.6 μg/mL and 0.25 μg/mL to 0.06 μg/mL respectively, moreover a reduced M. smegmatis growth in presence of RIF and STR compared with that of a control strain without LipF expression (p < 0.05 and p < 0.01) was shown.
Conclusions: LipF expression was associated with increased RIF and STR sensitivity in mycobacteria. Reduced LipF expression may contribute to the development of RIF and STR resistance in Mycobacterium species. Our findings provide information pertinent to understanding mycobacterial drug resistance mechanisms.
Keywords: LipF, lipases; Mycobacterium; Rifampicin-resistance; Streptomycin-resistance; Tuberculosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- WHO . Global Tuberculosis Report 2018. 2018.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

