The Helicobacter pylori Cag Type IV Secretion System
- PMID: 32451226
- PMCID: PMC7363556
- DOI: 10.1016/j.tim.2020.02.004
The Helicobacter pylori Cag Type IV Secretion System
Abstract
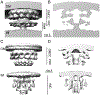
Colonization of the human stomach with Helicobacter pylori strains containing the cag pathogenicity island is a risk factor for development of gastric cancer. The cag pathogenicity island contains genes encoding a secreted effector protein (CagA) and components of a type IV secretion system (Cag T4SS). The molecular architecture of the H. pylori Cag T4SS is substantially more complex than that of prototype T4SSs in other bacterial species. In this review, we discuss recent discoveries pertaining to the structure and function of the Cag T4SS and its role in gastric cancer pathogenesis.
Keywords: bacterial nanomachines; bacterial protein secretion; cryo-electron microscopy; cryo-electron tomography; gastric adenocarcinoma; gastric cancer; macromolecular structures; peptic ulcer disease; type IV secretion system.
Published by Elsevier Ltd.
Figures

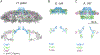
References
-
- Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-analysis. Gastroenterology doi: 10.1053/j.gastro.2017.04.022. - DOI - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. - PubMed
-
- Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Molecular Microbiology 28:37–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

