Extracellular serine controls epidermal stem cell fate and tumour initiation
- PMID: 32451440
- PMCID: PMC7343604
- DOI: 10.1038/s41556-020-0525-9
Extracellular serine controls epidermal stem cell fate and tumour initiation
Erratum in
-
Author Correction: Extracellular serine controls epidermal stem cell fate and tumour initiation.Nat Cell Biol. 2020 Nov;22(11):1396. doi: 10.1038/s41556-020-00600-x. Nat Cell Biol. 2020. PMID: 33046885
Abstract
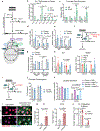
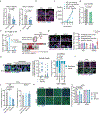
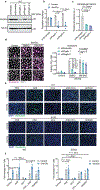
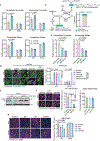
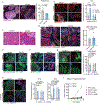
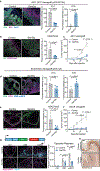
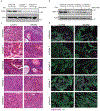
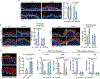
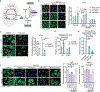
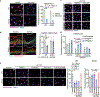
Tissue stem cells are the cell of origin for many malignancies. Metabolites regulate the balance between self-renewal and differentiation, but whether endogenous metabolic pathways or nutrient availability predispose stem cells towards transformation remains unknown. Here, we address this question in epidermal stem cells (EpdSCs), which are a cell of origin for squamous cell carcinoma. We find that oncogenic EpdSCs are serine auxotrophs whose growth and self-renewal require abundant exogenous serine. When extracellular serine is limited, EpdSCs activate de novo serine synthesis, which in turn stimulates α-ketoglutarate-dependent dioxygenases that remove the repressive histone modification H3K27me3 and activate differentiation programmes. Accordingly, serine starvation or enforced α-ketoglutarate production antagonizes squamous cell carcinoma growth. Conversely, blocking serine synthesis or repressing α-ketoglutarate-driven demethylation facilitates malignant progression. Together, these findings reveal that extracellular serine is a critical determinant of EpdSC fate and provide insight into how nutrient availability is integrated with stem cell fate decisions during tumour initiation.
Conflict of interest statement
Declaration of competing interests.
Authors declare no competing interests.
Figures







Comment in
-
Extracellular serine feeds cancer skin stem cells.Nat Rev Mol Cell Biol. 2020 Jul;21(7):359. doi: 10.1038/s41580-020-0261-9. Nat Rev Mol Cell Biol. 2020. PMID: 32488219 No abstract available.
-
Starve a cold, and perhaps a cancer.Nat Cell Biol. 2020 Jul;22(7):755-757. doi: 10.1038/s41556-020-0543-7. Nat Cell Biol. 2020. PMID: 32587343 Free PMC article.
-
Integrating serine with tumour initiation.Nat Rev Cancer. 2020 Aug;20(8):416. doi: 10.1038/s41568-020-0287-5. Nat Rev Cancer. 2020. PMID: 32612179 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

