Common germline variants of the human APOE gene modulate melanoma progression and survival
- PMID: 32451497
- PMCID: PMC8058866
- DOI: 10.1038/s41591-020-0879-3
Common germline variants of the human APOE gene modulate melanoma progression and survival
Abstract
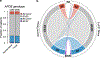
Common germline variants of the APOE gene are major risk modifiers of neurodegenerative and atherosclerotic diseases1-3, but their effect on cancer outcome is poorly defined. Here we report that, in a reversal of their effect on Alzheimer's disease, the APOE4 and APOE2 variants confer favorable and poor outcomes in melanoma, respectively. Mice expressing the human APOE4 allele exhibited reduced melanoma progression and metastasis relative to APOE2 mice. APOE4 mice exhibited enhanced anti-tumor immune activation relative to APOE2 mice, and T cell depletion experiments showed that the effect of APOE genotype on melanoma progression was mediated by altered anti-tumor immunity. Consistently, patients with melanoma carrying the APOE4 variant experienced improved survival in comparison to carriers of APOE2. Notably, APOE4 mice also showed improved outcomes under PD1 immune checkpoint blockade relative to APOE2 mice, and patients carrying APOE4 experienced improved anti-PD1 immunotherapy survival after progression on frontline regimens. Finally, enhancing APOE expression via pharmacologic activation of liver X receptors, previously shown to boost anti-tumor immunity4, exhibited therapeutic efficacy in APOE4 mice but not in APOE2 mice. These findings demonstrate that pre-existing hereditary genetics can impact progression and survival outcomes of a future malignancy and warrant prospective investigation of APOE genotype as a biomarker for melanoma outcome and therapeutic response.
Conflict of interest statement
Competing interests
S.F.T and B.N.O. are inventors on a US provisional patent application encompassing aspects of this work. S.F.T. is a co-founder, shareholder, and member of the scientific advisory board of Rgenix.
Figures
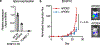



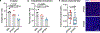
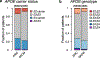






Comment in
-
Germline control of anti-tumour immunity.Nat Rev Cancer. 2020 Aug;20(8):414. doi: 10.1038/s41568-020-0282-x. Nat Rev Cancer. 2020. PMID: 32555350 No abstract available.
References
-
- Corder EH et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7, 180–4 (1994). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

