Genomic prediction of maize microphenotypes provides insights for optimizing selection and mining diversity
- PMID: 32452105
- PMCID: PMC7680549
- DOI: 10.1111/pbi.13420
Genomic prediction of maize microphenotypes provides insights for optimizing selection and mining diversity
Abstract
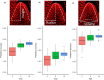
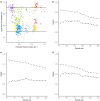
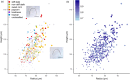
Effective evaluation of millions of crop genetic stocks is an essential component of exploiting genetic diversity to achieve global food security. By leveraging genomics and data analytics, genomic prediction is a promising strategy to efficiently explore the potential of these gene banks by starting with phenotyping a small designed subset. Reliable genomic predictions have enhanced selection of many macroscopic phenotypes in plants and animals. However, the use of genomicprediction strategies for analysis of microscopic phenotypes is limited. Here, we exploited the power of genomic prediction for eight maize traits related to the shoot apical meristem (SAM), the microscopic stem cell niche that generates all the above-ground organs of the plant. With 435 713 genomewide single-nucleotide polymorphisms (SNPs), we predicted SAM morphology traits for 2687 diverse maize inbreds based on a model trained from 369 inbreds. An empirical validation experiment with 488 inbreds obtained a prediction accuracy of 0.37-0.57 across eight traits. In addition, we show that a significantly higher prediction accuracy was achieved by leveraging the U value (upper bound for reliability) that quantifies the genomic relationships of the validation set with the training set. Our findings suggest that double selection considering both prediction and reliability can be implemented in choosing selection candidates for phenotyping when exploring new diversity is desired. In this case, individuals with less extreme predicted values and moderate reliability values can be considered. Our study expands the turbocharging gene banks via genomic prediction from the macrophenotypes into the microphenotypic space.
Keywords: genetic diversity; genomic selection; genomics; maize; plant breeding; shoot apical meristem.
© 2020 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bernardo, R. and Yu, J. (2007) Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47, 1082–1090.
-
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. - PubMed
-
- de los Campos, G. , Gianola, D. and Allison, D.B. (2010) Predicting genetic predisposition in humans: the promise of whole‐genome markers. Nat. Rev. Genet. 11, 880–886. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

