Mesenchymal stromal cells induce distinct myeloid-derived suppressor cells in inflammation
- PMID: 32453713
- PMCID: PMC7406289
- DOI: 10.1172/jci.insight.136059
Mesenchymal stromal cells induce distinct myeloid-derived suppressor cells in inflammation
Abstract
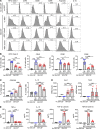
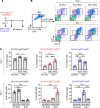
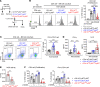
Mesenchymal stem/stromal cells (MSCs) regulate immunity through myeloid-derived suppressor cells (MDSCs), which are a heterogeneous population of immature myeloid cells with phenotypic and functional diversity. Herein, we identified a distinct subset of MDSCs induced by MSCs in the BM under inflammatory conditions. MSCs directed the differentiation of Ly6Glo BM cells from CD11bhiLy6Chi cells to CD11bmidLy6Cmid cells both in cell contact-independent and -dependent manners upon GM-CSF stimulation in vitro and in mice with experimental autoimmune uveoretinitis (EAU). RNA-Seq indicated that MSC-induced CD11bmidLy6CmidLy6Glo cells had a distinct transcriptome profile from CD11bhiLy6ChiLy6Glo cells. Phenotypic, molecular, and functional analyses showed that CD11bmidLy6CmidLy6Glo cells differed from CD11bhiLy6ChiLy6Glo cells by low expression of MHC class II and costimulatory molecules and proinflammatory cytokines, high production of immunoregulatory molecules, lack of change in response to LPS, and inhibition of T cell proliferation and activation. Consequently, adoptive transfer of MSC-induced CD11bmidLy6CmidLy6Glo cells significantly attenuated the development of EAU in mice. Further mechanistic study revealed that suppression of prostaglandin E2 (PGE2) and HGF secretion in MSCs by siRNA transfection partially reversed the effects of MSCs on MDSC differentiation. Altogether, data demonstrate that MSCs drive the differentiation of BM cells toward CD11bmidLy6CmidLy6Glo MDSCs, in part through HGF and COX-2/PGE2, leading to resolution of ocular autoimmune inflammation.
Keywords: Adult stem cells; Autoimmune diseases; Autoimmunity; Immunology; Stem cell transplantation.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

