Dysfunction of the ciliary ARMC9/TOGARAM1 protein module causes Joubert syndrome
- PMID: 32453716
- PMCID: PMC7410078
- DOI: 10.1172/JCI131656
Dysfunction of the ciliary ARMC9/TOGARAM1 protein module causes Joubert syndrome
Abstract
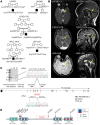
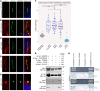
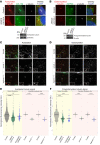
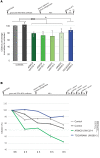
Joubert syndrome (JBTS) is a recessive neurodevelopmental ciliopathy characterized by a pathognomonic hindbrain malformation. All known JBTS genes encode proteins involved in the structure or function of primary cilia, ubiquitous antenna-like organelles essential for cellular signal transduction. Here, we used the recently identified JBTS-associated protein armadillo repeat motif-containing 9 (ARMC9) in tandem-affinity purification and yeast 2-hybrid screens to identify a ciliary module whose dysfunction underlies JBTS. In addition to the known JBTS-associated proteins CEP104 and CSPP1, we identified coiled-coil domain containing 66 (CCDC66) and TOG array regulator of axonemal microtubules 1 (TOGARAM1) as ARMC9 interaction partners. We found that TOGARAM1 variants cause JBTS and disrupt TOGARAM1 interaction with ARMC9. Using a combination of protein interaction analyses, characterization of patient-derived fibroblasts, and analysis of CRISPR/Cas9-engineered zebrafish and hTERT-RPE1 cells, we demonstrated that dysfunction of ARMC9 or TOGARAM1 resulted in short cilia with decreased axonemal acetylation and polyglutamylation, but relatively intact transition zone function. Aberrant serum-induced ciliary resorption and cold-induced depolymerization in ARMC9 and TOGARAM1 patient cell lines suggest a role for this new JBTS-associated protein module in ciliary stability.
Keywords: Genetic diseases; Genetics; Proteomics.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

