Gene expression regulation by the Chromodomain helicase DNA-binding protein 9 (CHD9) chromatin remodeler is dispensable for murine development
- PMID: 32453735
- PMCID: PMC7250415
- DOI: 10.1371/journal.pone.0233394
Gene expression regulation by the Chromodomain helicase DNA-binding protein 9 (CHD9) chromatin remodeler is dispensable for murine development
Abstract
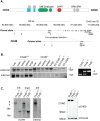
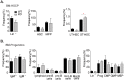
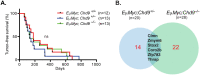
Chromodomain helicase DNA-binding (CHD) chromatin remodelers regulate transcription and DNA repair. They govern cell-fate decisions during embryonic development and are often deregulated in human pathologies. Chd1-8 show upon germline disruption pronounced, often developmental lethal phenotypes. Here we show that contrary to Chd1-8 disruption, Chd9-/-animals are viable, fertile and display no developmental defects or disease predisposition. Germline deletion of Chd9 only moderately affects gene expression in tissues and derived cells, whereas acute depletion in human cancer cells elicits more robust changes suggesting that CHD9 is a highly context-dependent chromatin regulator that, surprisingly, is dispensable for mouse development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

