Glucose transporter 10 modulates adipogenesis via an ascorbic acid-mediated pathway to protect mice against diet-induced metabolic dysregulation
- PMID: 32453789
- PMCID: PMC7274451
- DOI: 10.1371/journal.pgen.1008823
Glucose transporter 10 modulates adipogenesis via an ascorbic acid-mediated pathway to protect mice against diet-induced metabolic dysregulation
Abstract
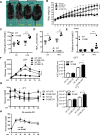
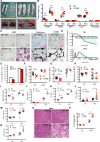
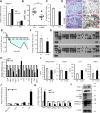
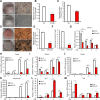
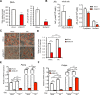
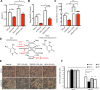
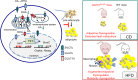
The development of type 2 diabetes mellitus (T2DM) depends on interactions between genetic and environmental factors, and a better understanding of gene-diet interactions in T2DM will be useful for disease prediction and prevention. Ascorbic acid has been proposed to reduce the risk of T2DM. However, the links between ascorbic acid and metabolic consequences are not fully understood. Here, we report that glucose transporter 10 (GLUT10) maintains intracellular levels of ascorbic acid to promote adipogenesis, white adipose tissue (WAT) development and protect mice from high-fat diet (HFD)-induced metabolic dysregulation. We found genetic polymorphisms in SLC2A10 locus are suggestively associated with a T2DM intermediate phenotype in non-diabetic Han Taiwanese. Additionally, mice carrying an orthologous human Glut10G128E variant (Glut10G128E mice) with compromised GLUT10 function have reduced adipogenesis, reduced WAT development and increased susceptibility to HFD-induced metabolic dysregulation. We further demonstrate that GLUT10 is highly expressed in preadipocytes, where it regulates intracellular ascorbic acid levels and adipogenesis. In this context, GLUT10 increases ascorbic acid-dependent DNA demethylation and the expression of key adipogenic genes, Cebpa and Pparg. Together, our data show GLUT10 regulates adipogenesis via ascorbic acid-dependent DNA demethylation to benefit proper WAT development and protect mice against HFD-induced metabolic dysregulation. Our findings suggest that SLC2A10 may be an important HFD-associated susceptibility locus for T2DM.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, South Asian Type 2 Diabetes C, Mexican American Type 2 Diabetes C, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples C, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–44. 10.1038/ng.2897 - DOI - PMC - PubMed
-
- Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 2014;111(36):13127–32. Epub 2014/08/27. 10.1073/pnas.1410428111 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

