Pangenome Graphs
- PMID: 32453966
- PMCID: PMC8006571
- DOI: 10.1146/annurev-genom-120219-080406
Pangenome Graphs
Abstract
Low-cost whole-genome assembly has enabled the collection of haplotype-resolved pangenomes for numerous organisms. In turn, this technological change is encouraging the development of methods that can precisely address the sequence and variation described in large collections of related genomes. These approaches often use graphical models of the pangenome to support algorithms for sequence alignment, visualization, functional genomics, and association studies. The additional information provided to these methods by the pangenome allows them to achieve superior performance on a variety of bioinformatic tasks, including read alignment, variant calling, and genotyping. Pangenome graphs stand to become a ubiquitous tool in genomics. Although it is unclear whether they will replace linearreference genomes, their ability to harmoniously relate multiple sequence and coordinate systems will make them useful irrespective of which pangenomic models become most common in the future.
Keywords: genome graph; pangenome; variation graph.
Conflict of interest statement
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Figures
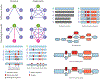
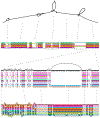

References
-
- Amir A, Lewenstein M, Lewenstein N. 1997. Pattern matching in hypertext. In Lecture Notes in Computer Science. Springer Berlin Heidelberg, 160–173
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

