HuRdling Senescence: HuR Breaks BRAF-Induced Senescence in Melanocytes and Supports Melanoma Growth
- PMID: 32455577
- PMCID: PMC7281285
- DOI: 10.3390/cancers12051299
HuRdling Senescence: HuR Breaks BRAF-Induced Senescence in Melanocytes and Supports Melanoma Growth
Abstract
In addition to genetic changes, post-transcriptional events strongly contribute to the progression of malignant tumors. The RNA-binding protein HuR (ELAVL1) is able to bind and stabilize a large group of target mRNAs, which contain AU-rich elements (ARE) in their 3'-untranslated region. We found HuR to be upregulated in malignant melanoma in vitro and in vivo, significantly correlating with progression in vivo. Additionally, we could show that miR-194-5p can regulate HuR expression level. HuR knockdown in melanoma cells led to the suppression of proliferation and the induction of cellular senescence. Interestingly, HuR overexpression was sufficient to inhibit senescence in BRAFV600E-expressing melanocytes and to force their growth. Here, MITF (Microphthalmia-associated transcription factor), a key player in suppressing senescence and an ARE containing transcript, is positively regulated by HuR. Our results show for the first time that the overexpression of HuR is an important part of the regulatory pathway in the development of malignant melanoma and functions as a switch to overcome oncogene-induced senescence and to support melanoma formation. These newly defined alterations may provide possibilities for innovative therapeutic approaches.
Keywords: HuR; MITF; Microphthalmia-associated transcription factor; malignant melanoma; oncogene induced senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


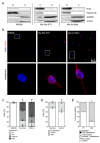
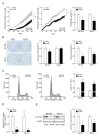
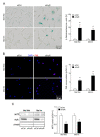

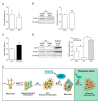
References
-
- Bernatchez C., Cooper Z.A., Wargo J.A., Hwu P., Lizee G. Novel Treatments in Development for Melanoma. Cancer Treat. Res. 2016;167:371–416. - PubMed
-
- Long G.V., Menzies A.M., Nagrial A.M., Haydu L.E., Hamilton A.L., Mann G.J., Hughes T.M., Thompson J.F., Scolyer R.A., Kefford R.F. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J. Clin. Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

