Distinct Tumor Microenvironments Are a Defining Feature of Strain-Specific CRISPR/Cas9-Induced MPNSTs
- PMID: 32456131
- PMCID: PMC7288323
- DOI: 10.3390/genes11050583
Distinct Tumor Microenvironments Are a Defining Feature of Strain-Specific CRISPR/Cas9-Induced MPNSTs
Abstract
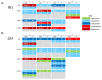
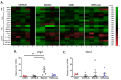
The tumor microenvironment plays important roles in cancer biology, but genetic backgrounds of mouse models can complicate interpretation of tumor phenotypes. A deeper understanding of strain-dependent influences on the tumor microenvironment of genetically-identical tumors is critical to exploring genotype-phenotype relationships, but these interactions can be difficult to identify using traditional Cre/loxP approaches. Here, we use somatic CRISPR/Cas9 tumorigenesis approaches to determine the impact of mouse background on the biology of genetically-identical malignant peripheral nerve sheath tumors (MPNSTs) in four commonly-used inbred strains. To our knowledge, this is the first study to systematically evaluate the impact of host strain on CRISPR/Cas9-generated mouse models. Our data identify multiple strain-dependent phenotypes, including changes in tumor onset and the immune microenvironment. While BALB/c mice develop MPNSTs earlier than other strains, similar tumor onset is observed in C57BL/6, 129X1 and 129/SvJae mice. Indel pattern analysis demonstrates that indel frequency, type and size are similar across all genetic backgrounds. Gene expression and IHC analysis identify multiple strain-dependent differences in CD4+ T cell infiltration and myeloid cell populations, including M2 macrophages and mast cells. These data highlight important strain-specific phenotypes of genomically-matched MPNSTs that have implications for the design of future studies using similar in vivo gene editing approaches.
Keywords: CRISPR/Cas9; MPNST; mouse models; sarcoma; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kuperwasser C., Hurlbut G.D., Kittrell F.S., Dickinson E.S., Laucirica R., Medina D., Naber S.P., Jerry D.J. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am. J. Pathol. 2000;157:2151–2159. doi: 10.1016/S0002-9440(10)64853-5. - DOI - PMC - PubMed
-
- Blackburn A.C., Hill L.Z., Roberts A.L., Wang J., Aud D., Jung J., Nikolcheva T., Allard J., Peltz G., Otis C.N., et al. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. Am. J. Pathol. 2007;170:2030–2041. doi: 10.2353/ajpath.2007.060512. - DOI - PMC - PubMed
-
- Brandt L.P., Albers J., Hejhal T., Pfundstein S., Gonçalves A.F., Catalano A., Wild P.J., Frew I.J. Mouse genetic background influences whether HrasG12V expression plus Cdkn2a knockdown causes angiosarcoma or undifferentiated pleomorphic sarcoma. Oncotarget. 2018;9:19753–19766. doi: 10.18632/oncotarget.24831. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

