Genome-Wide Identification and Evolutionary Analysis of NBS-LRR Genes From Dioscorea rotundata
- PMID: 32457809
- PMCID: PMC7224235
- DOI: 10.3389/fgene.2020.00484
Genome-Wide Identification and Evolutionary Analysis of NBS-LRR Genes From Dioscorea rotundata
Abstract
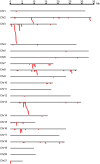
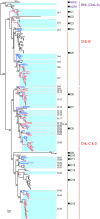
Dioscorea rotundata is an important food crop that is mainly cultivated in subtropical regions of the world. D. rotundata is frequently infected by various pathogens during its lifespan, which results in a substantial economic loss in terms of yield and quality. The disease resistance gene (R gene) profile of D. rotundata is largely unknown, which has greatly hampered molecular study of disease resistance in this species. Nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes are the largest group of plant R genes, and they play important roles in plant defense responses to various pathogens. In this study, 167 NBS-LRR genes were identified from the D. rotundata genome. Subsequently, one gene was assigned to the resistance to powdery mildew8 (RPW8)-NBS-LRR (RNL) subclass and the other 166 genes to the coiled coil (CC)-NBS-LRR (CNL) subclass. None of the Toll/interleukin-1 receptor (TIR)-NBS-LRR (TNL) genes were detected in the genome. Among them, 124 genes are located in 25 multigene clusters and 43 genes are singletons. Tandem duplication serves as the major force for the cluster arrangement of NBS-LRR genes. Segmental duplication was detected for 18 NBS-LRR genes, although no whole-genome duplication has been documented for the species. Phylogenetic analysis revealed that D. rotundata NBS-LRR genes share 15 ancestral lineages with Arabidopsis thaliana genes. The NBS-LRR gene number increased by more than a factor of 10 during D. rotundata evolution. A conservatively evolved ancestral lineage was identified from D. rotundata, which is orthologs to the Arabidopsis RPM1 gene. Transcriptome analysis for four different tissues of D. rotundata revealed a low expression of most NBS-LRR genes, with the tuber and leaf displaying a relatively high NBS-LRR gene expression than the stem and flower. Overall, this study provides a complete set of NBS-LRR genes for D. rotundata, which may serve as a fundamental resource for mining functional NBS-LRR genes against various pathogens.
Keywords: Dioscorea rotundata; NBS-LRR genes; R gene; genome-wide analysis; pathogen defense.
Copyright © 2020 Zhang, Chen, Sun, Wang, Yin, Liu, Sun and Hang.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

